The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation
- PMID: 20360945
- PMCID: PMC2847742
- DOI: 10.1593/neo.10144
The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation
Abstract
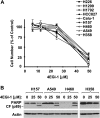
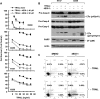
The small molecule 4EGI-1 was identified as an inhibitor of cap-dependent translation initiation owing to its disruption of the eIF4E/eIF4G association through binding to eIF4E. 4EGI-1 exhibits growth-inhibitory and apoptosis-inducing activity in cancer cells; thus, we were interested in its therapeutic efficacy in human lung cancer cells. 4EGI-1, as a single agent, inhibited the growth and induced apoptosis of human lung cancer cells.When combined with the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), enhanced apoptosis-induced activity was observed. As expected, 4EGI-1 inhibited eIF4E/eIF4G interaction and reduced the levels of cyclin D1 and hypoxia-inducing factor-1alpha (HIF-1alpha), both of which are regulated by a cap-dependent translation mechanism. Moreover, 4EGI-1 induced CCAAT/enhancer-binding protein homologous protein-dependent DR5 expression and ubiquitin/proteasome- mediated degradation of cellular FLICE-inhibitory protein (c-FLIP). Small interfering RNA-mediated blockade of DR5 induction or enforced expression of c-FLIP abrogated 4EGI-1's ability to enhance TRAIL-induced apoptosis, indicating that both DR5 induction and c-FLIP down-regulation contribute to enhancement of TRAIL-induced apoptosis by 4EGI-1. However, inhibition of eIF4E/eIF4G interaction by knockdown of eIF4E effectively reduced the levels of cyclin D1 and HIF-1alpha but failed to induce DR5 expression, downregulate c-FLIP levels, or augment TRAIL-induced apoptosis. These results collectively suggest that 4EGI-1 augments TRAIL-induced apoptosis through induction of DR5 and down-regulation of c-FLIP, independent of inhibition of cap-dependent protein translation.
Figures




Similar articles
-
CCAAT/enhancer binding protein homologous protein-dependent death receptor 5 induction and ubiquitin/proteasome-mediated cellular FLICE-inhibitory protein down-regulation contribute to enhancement of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by dimethyl-celecoxib in human non small-cell lung cancer cells.Mol Pharmacol. 2007 Nov;72(5):1269-79. doi: 10.1124/mol.107.037465. Epub 2007 Aug 7. Mol Pharmacol. 2007. PMID: 17684158
-
The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells.Cancer Res. 2007 May 15;67(10):4981-8. doi: 10.1158/0008-5472.CAN-06-4274. Cancer Res. 2007. PMID: 17510429
-
Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G.Cell. 2007 Jan 26;128(2):257-67. doi: 10.1016/j.cell.2006.11.046. Cell. 2007. PMID: 17254965
-
Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL.J Cell Physiol. 2018 Oct;233(10):6470-6485. doi: 10.1002/jcp.26585. Epub 2018 May 9. J Cell Physiol. 2018. PMID: 29741767 Review.
-
c-FLIP, a master anti-apoptotic regulator.Exp Oncol. 2012 Oct;34(3):176-84. Exp Oncol. 2012. PMID: 23070002 Free PMC article. Review.
Cited by
-
Synthesis of Alkyl/Aryloxymethyl Derivatives of 1,2,4-Triazole-3-Carboxamides and Their Biological Activities.Molecules. 2024 Oct 11;29(20):4808. doi: 10.3390/molecules29204808. Molecules. 2024. PMID: 39459177 Free PMC article.
-
Overcoming intratumor heterogeneity of polygenic cancer drug resistance with improved biomarker integration.Neoplasia. 2012 Dec;14(12):1278-89. doi: 10.1593/neo.122096. Neoplasia. 2012. PMID: 23308059 Free PMC article.
-
Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer.Cancer Biol Ther. 2012 Mar;13(5):272-80. doi: 10.4161/cbt.18923. Epub 2012 Mar 1. Cancer Biol Ther. 2012. PMID: 22236867 Free PMC article.
-
The synergistic inhibition of breast cancer proliferation by combined treatment with 4EGI-1 and MK2206.Cell Cycle. 2015;14(2):232-42. doi: 10.4161/15384101.2014.977096. Cell Cycle. 2015. PMID: 25607647 Free PMC article.
-
Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1.J Virol. 2011 Jan;85(2):853-64. doi: 10.1128/JVI.01873-10. Epub 2010 Nov 10. J Virol. 2011. PMID: 21068241 Free PMC article.
References
-
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol. 2004;11:503–511. - PubMed
-
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. - PubMed
-
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. - PubMed
-
- Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. - PubMed
-
- De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
