An accessory to the 'Trinity': SR-As are essential pathogen sensors of extracellular dsRNA, mediating entry and leading to subsequent type I IFN responses
- PMID: 20360967
- PMCID: PMC2847946
- DOI: 10.1371/journal.ppat.1000829
An accessory to the 'Trinity': SR-As are essential pathogen sensors of extracellular dsRNA, mediating entry and leading to subsequent type I IFN responses
Abstract
Extracellular RNA is becoming increasingly recognized as a signaling molecule. Virally derived double stranded (ds)RNA released into the extracellular space during virus induced cell lysis acts as a powerful inducer of classical type I interferon (IFN) responses; however, the receptor that mediates this response has not been identified. Class A scavenger receptors (SR-As) are likely candidates due to their cell surface expression and ability to bind nucleic acids. In this study, we investigated a possible role for SR-As in mediating type I IFN responses induced by extracellular dsRNA in fibroblasts, a predominant producer of IFNbeta. Fibroblasts were found to express functional SR-As, even SR-A species thought to be macrophage specific. SR-A specific competitive ligands significantly blocked extracellular dsRNA binding, entry and subsequent interferon stimulated gene (ISG) induction. Candidate SR-As were systematically investigated using RNAi and the most dramatic inhibition in responses was observed when all candidate SR-As were knocked down in unison. Partial inhibition of dsRNA induced antiviral responses was observed in vivo in SR-AI/II(-/-) mice compared with WT controls. The role of SR-As in mediating extracellular dsRNA entry and subsequent induced antiviral responses was observed in both murine and human fibroblasts. SR-As appear to function as 'carriers', facilitating dsRNA entry and delivery to the established dsRNA sensing receptors, specifically TLR3, RIGI and MDA-5. Identifying SR-As as gatekeepers of the cell, mediating innate antiviral responses, represents a novel function for this receptor family and provides insight into how cells recognize danger signals associated with lytic virus infections. Furthermore, the implications of a cell surface receptor capable of recognizing extracellular RNA may exceed beyond viral immunity to mediating other important innate immune functions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

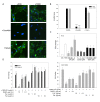

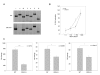
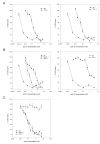

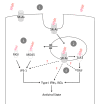
Similar articles
-
Class A scavenger receptors mediate extracellular dsRNA sensing, leading to downstream antiviral gene expression in a novel American toad cell line, BufoTad.Dev Comp Immunol. 2019 Mar;92:140-149. doi: 10.1016/j.dci.2018.11.012. Epub 2018 Nov 16. Dev Comp Immunol. 2019. PMID: 30452932
-
Class A Scavenger Receptor-Mediated Double-Stranded RNA Internalization Is Independent of Innate Antiviral Signaling and Does Not Require Phosphatidylinositol 3-Kinase Activity.J Immunol. 2015 Oct 15;195(8):3858-65. doi: 10.4049/jimmunol.1501028. Epub 2015 Sep 11. J Immunol. 2015. PMID: 26363049 Free PMC article.
-
Understanding Viral dsRNA-Mediated Innate Immune Responses at the Cellular Level Using a Rainbow Trout Model.Front Immunol. 2018 Apr 23;9:829. doi: 10.3389/fimmu.2018.00829. eCollection 2018. Front Immunol. 2018. PMID: 29740439 Free PMC article.
-
Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing.Immunol Rev. 2009 Jan;227(1):44-53. doi: 10.1111/j.1600-065X.2008.00723.x. Immunol Rev. 2009. PMID: 19120474 Review.
-
Beyond dsRNA: Toll-like receptor 3 signalling in RNA-induced immune responses.Biochem J. 2014 Mar 1;458(2):195-201. doi: 10.1042/BJ20131492. Biochem J. 2014. PMID: 24524192 Review.
Cited by
-
Emerging complexity and new roles for the RIG-I-like receptors in innate antiviral immunity.Virol Sin. 2015 Jun;30(3):163-73. doi: 10.1007/s12250-015-3604-5. Epub 2015 May 20. Virol Sin. 2015. PMID: 25997992 Free PMC article. Review.
-
Discovery and Use of Long dsRNA Mediated RNA Interference to Stimulate Antiviral Protection in Interferon Competent Mammalian Cells.Front Immunol. 2022 May 6;13:859749. doi: 10.3389/fimmu.2022.859749. eCollection 2022. Front Immunol. 2022. PMID: 35603190 Free PMC article.
-
Distinct roles for the NF-kappa B RelA subunit during antiviral innate immune responses.J Virol. 2011 Mar;85(6):2599-610. doi: 10.1128/JVI.02213-10. Epub 2011 Jan 5. J Virol. 2011. PMID: 21209118 Free PMC article.
-
Scavenger receptor class A member 3 (SCARA3) in disease progression and therapy resistance in multiple myeloma.Leuk Res. 2013 Aug;37(8):963-9. doi: 10.1016/j.leukres.2013.03.004. Epub 2013 Mar 26. Leuk Res. 2013. PMID: 23537707 Free PMC article.
-
Expression of scavenger receptor-AI promotes alternative activation of murine macrophages to limit hepatic inflammation and fibrosis.Hepatology. 2017 Jan;65(1):32-43. doi: 10.1002/hep.28873. Epub 2016 Nov 22. Hepatology. 2017. PMID: 27770558 Free PMC article.
References
-
- Creagh EM, O'Neill LAJ. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. - PubMed
-
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF- κB by Toll-like receptor 3. Nature. 2001;413:732–738. - PubMed
-
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

