Rapid evolution of pandemic noroviruses of the GII.4 lineage
- PMID: 20360972
- PMCID: PMC2847951
- DOI: 10.1371/journal.ppat.1000831
Rapid evolution of pandemic noroviruses of the GII.4 lineage
Erratum in
- PLoS Pathog. 2010;6(4) doi.org/10.1371/annotation/19042899-9f1b-4ccc-b13e-2a8faf19421b. White, Peter A [added]
Abstract
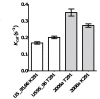
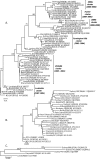
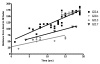
Over the last fifteen years there have been five pandemics of norovirus (NoV) associated gastroenteritis, and the period of stasis between each pandemic has been progressively shortening. NoV is classified into five genogroups, which can be further classified into 25 or more different human NoV genotypes; however, only one, genogroup II genotype 4 (GII.4), is associated with pandemics. Hence, GII.4 viruses have both a higher frequency in the host population and greater epidemiological fitness. The aim of this study was to investigate if the accuracy and rate of replication are contributing to the increased epidemiological fitness of the GII.4 strains. The replication and mutation rates were determined using in vitro RNA dependent RNA polymerase (RdRp) assays, and rates of evolution were determined by bioinformatics. GII.4 strains were compared to the second most reported genotype, recombinant GII.b/GII.3, the rarely detected GII.3 and GII.7 and as a control, hepatitis C virus (HCV). The predominant GII.4 strains had a higher mutation rate and rate of evolution compared to the less frequently detected GII.b, GII.3 and GII.7 strains. Furthermore, the GII.4 lineage had on average a 1.7-fold higher rate of evolution within the capsid sequence and a greater number of non-synonymous changes compared to other NoVs, supporting the theory that it is undergoing antigenic drift at a faster rate. Interestingly, the non-synonymous mutations for all three NoV genotypes were localised to common structural residues in the capsid, indicating that these sites are likely to be under immune selection. This study supports the hypothesis that the ability of the virus to generate genetic diversity is vital for viral fitness.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
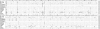
References
-
- Estes MK, Prasad BV, Atmar RL. Noroviruses everywhere: has something changed? Curr Opin Infect Dis. 2006;19:467–474. - PubMed
-
- Manrubia SC, Escarmis C, Domingo E, Lazaro E. High mutation rates, bottlenecks, and robustness of RNA viral quasispecies. Gene. 2005;347:273–282. - PubMed
-
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, et al. Mechanisms of GII.4 Norovirus Persistence in Human Populations. PLoS Med. 2008;5:e31. doi: 10.1371/journal.pmed.0050031. - DOI - PMC - PubMed
-
- Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, et al. Basic concepts in RNA virus evolution. Faseb J. 1996;10:859–864. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

