Unraveling polyketide synthesis in members of the genus Aspergillus
- PMID: 20361326
- PMCID: PMC3110678
- DOI: 10.1007/s00253-010-2525-3
Unraveling polyketide synthesis in members of the genus Aspergillus
Abstract
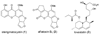
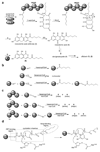
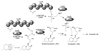
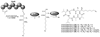
Aspergillus species have the ability to produce a wide range of secondary metabolites including polyketides that are generated by multi-domain polyketide synthases (PKSs). Recent biochemical studies using dissected single or multiple domains from PKSs have provided deep insight into how these PKSs control the structural outcome. Moreover, the recent genome sequencing of several species has greatly facilitated the understanding of the biosynthetic pathways for these secondary metabolites. In this review, we will highlight the current knowledge regarding polyketide biosynthesis in Aspergillus based on the domain architecture of non-reducing, highly reducing, and partially reducing PKSs, and PKS-non-ribosomal peptide synthetases.
Figures






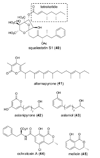


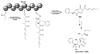
References
-
- Abe Y, Suzuki T, Ono C, Iwamoto K, Hosobuchi M, Yoshikawa H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol Genet Genomics. 2002;267:636–646. - PubMed
-
- Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. - PubMed
-
- Awakawa T, Yokota K, Funa N, Doi F, Mori N, Watanabe H, Horinouchi S. Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem Biol. 2009;16:613–623. - PubMed
-
- Bacha N, Atoui A, Mathieu F, Liboz T, Lebrihi A. Aspergillus westerdijkiae polyketide synthase gene “aoks1” is involved in the biosynthesis of ochratoxin A. Fungal Genet Biol. 2009a;46:77–84. - PubMed
-
- Bacha N, Dao HP, Atoui A, Mathieu F, O'Callaghan J, Puel O, Liboz T, Dobson AD, Lebrihi A. Cloning and characterization of novel methylsalicylic acid synthase gene involved in the biosynthesis of isoasperlactone and asperlactone in Aspergillus westerdijkiae. Fungal Genet Biol. 2009b;46:742–749. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

