An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus
- PMID: 20362316
- PMCID: PMC2873121
- DOI: 10.1016/j.virol.2010.03.013
An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus
Abstract
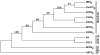
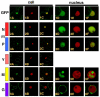
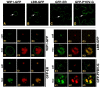
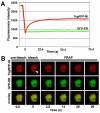
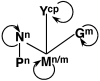
The genome of Potato yellow dwarf virus (PYDV; Nucleorhabdovirus type species) was determined to be 12,875 nucleotides (nt). The antigenome is organized into seven open reading frames (ORFs) ordered 3'-N-X-P-Y-M-G-L-5', which likely encode the nucleocapsid, phospho, movement, matrix, glyco and RNA-dependent RNA polymerase proteins, respectively, except for X, which is of unknown function. The ORFs are flanked by a 3' leader RNA of 149 nt and a 5' trailer RNA of 97 nt, and are separated by conserved intergenic junctions. Phylogenetic analyses indicated that PYDV is closely related to other leafhopper-transmitted rhabdoviruses. Functional protein assays were used to determine the subcellular localization of PYDV proteins. Surprisingly, the M protein was able to induce the intranuclear accumulation of the inner nuclear membrane in the absence of any other viral protein. Finally, bimolecular fluorescence complementation was used to generate the most comprehensive protein interaction map for a plant-adapted rhabdovirus to date.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures



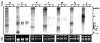


References
-
- Adam G, Gaedigk K. Inhibition of Potato Yellow Dwarf Virus infection in vector cell monolayers by lysosomotropic agents. J. Gen. Virol. 1986;67:2775–2780.
-
- Adam G, Hsu HT. Comparison of structural proteins from two Potato yellow dwarf viruses. J. Gen. Virol. 1984;65:991–994.
-
- Ammar el-D., Tsai CW, Whitfield AE, Redinbaugh MG, Hogenhout SA. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 2009;54:447–468. - PubMed
-
- Barr JN, Whelan SP, Wertz GW. Transcriptional control of the RNA-dependent RNA polymerase of Vesicular stomatitis virus. Biochem. Biophys. Acta. 2002;1577:337–353. - PubMed
-
- Barrus MF, Chupp CC. Yellow dwarf of potatoes. Phytopathology. 1922;12:122–133.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

