RNA traffic control of chromatin complexes
- PMID: 20362426
- PMCID: PMC2895502
- DOI: 10.1016/j.gde.2010.03.003
RNA traffic control of chromatin complexes
Abstract
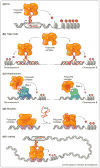
It is widely accepted that the genome is regulated by histone modifications that induce epigenetic changes on the genome. However, it is still not understood how ubiquitously expressed chromatin modifying complexes are 'guided' to specific genomic sites to induce intricate patterns of epigenetic modifications. Previously believed to represent 'genome junk', it is now becoming increasingly clear that large non-coding RNAs associate with chromatin modifying complexes. Here we explore an intriguing hypothesis that large non-coding RNA molecules might represent a molecular trafficking system that modulates chromatin modifying complexes to establish specific epigenetic landscapes.
Published by Elsevier Ltd.
Figures

References
-
- Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. - PubMed
-
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. - PubMed
-
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a shortrepeat RNA to the mouse X chromosome. Science. 2008;322:750–756. The study was the first to determine that RepA directly binds to the PRC2 chromatin modifying complex (likely through Ezh2) and is required for proper H3K27me3 localization on the inactive X chromosome. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

