The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase
- PMID: 20363755
- PMCID: PMC2878546
- DOI: 10.1074/jbc.M110.114785
The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase
Abstract
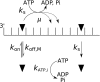
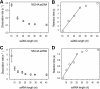
Hepatitis C virus (HCV) NS3 protein has two enzymatic activities of helicase and protease that are essential for viral replication. The helicase separates the strands of DNA and RNA duplexes using the energy from ATP hydrolysis. To understand how ATP hydrolysis is coupled to helicase movement, we measured the single turnover helicase translocation-dissociation kinetics and the pre-steady-state P(i) release kinetics on single-stranded RNA and DNA substrates of different lengths. The parameters of stepping were determined from global fitting of the two types of kinetic measurements into a computational model that describes translocation as a sequence of coupled hydrolysis-stepping reactions. Our results show that the HCV helicase moves with a faster rate on single stranded RNA than on DNA. The HCV helicase steps on the RNA or DNA one nucleotide at a time, and due to imperfect coupling, not every ATP hydrolysis event produces a successful step. Comparison of the helicase domain (NS3h) with the protease-helicase (NS3-4A) shows that the most significant contribution of the protease domain is to improve the translocation stepping efficiency of the helicase. Whereas for NS3h, only 20% of the hydrolysis events result in translocation, the coupling for NS3-4A is near-perfect 93%. The presence of the protease domain also significantly reduces the stepping rate, but it doubles the processivity. These effects of the protease domain on the helicase can be explained by an improved allosteric cross-talk between the ATP- and nucleic acid-binding sites achieved by the overall stabilization of the helicase domain structure.
Figures



Similar articles
-
Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites.Biochemistry. 2002 Aug 13;41(32):10332-42. doi: 10.1021/bi026082g. Biochemistry. 2002. PMID: 12162749
-
Investigation of translocation, DNA unwinding, and protein displacement by NS3h, the helicase domain from the hepatitis C virus helicase.Biochemistry. 2010 Mar 16;49(10):2097-109. doi: 10.1021/bi901977k. Biochemistry. 2010. PMID: 20108974 Free PMC article.
-
Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus.J Biol Chem. 2006 Feb 10;281(6):3528-35. doi: 10.1074/jbc.M512100200. Epub 2005 Nov 22. J Biol Chem. 2006. PMID: 16306038
-
Structure and function of hepatitis C virus NS3 helicase.Curr Top Microbiol Immunol. 2000;242:171-96. doi: 10.1007/978-3-642-59605-6_9. Curr Top Microbiol Immunol. 2000. PMID: 10592661 Review.
-
HCV Helicase: Structure, Function, and Inhibition.In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 7. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 7. PMID: 21250378 Free Books & Documents. Review.
Cited by
-
Stepwise nucleosome translocation by RSC remodeling complexes.Elife. 2016 Feb 19;5:e10051. doi: 10.7554/eLife.10051. Elife. 2016. PMID: 26895087 Free PMC article.
-
RecQ helicase translocates along single-stranded DNA with a moderate processivity and tight mechanochemical coupling.Proc Natl Acad Sci U S A. 2012 Jun 19;109(25):9804-9. doi: 10.1073/pnas.1114468109. Epub 2012 Jun 4. Proc Natl Acad Sci U S A. 2012. PMID: 22665805 Free PMC article.
-
Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis.J Mol Biol. 2012 Apr 20;418(1-2):32-46. doi: 10.1016/j.jmb.2012.02.013. Epub 2012 Feb 14. J Mol Biol. 2012. PMID: 22342931 Free PMC article.
-
Structure-based simulations of the translocation mechanism of the hepatitis C virus NS3 helicase along single-stranded nucleic acid.Biophys J. 2012 Sep 19;103(6):1343-53. doi: 10.1016/j.bpj.2012.08.026. Biophys J. 2012. PMID: 22995507 Free PMC article.
-
HCV Induces Telomerase Reverse Transcriptase, Increases Its Catalytic Activity, and Promotes Caspase Degradation in Infected Human Hepatocytes.PLoS One. 2017 Jan 5;12(1):e0166853. doi: 10.1371/journal.pone.0166853. eCollection 2017. PLoS One. 2017. PMID: 28056029 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

