CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes
- PMID: 20363758
- PMCID: PMC2885212
- DOI: 10.1074/jbc.M110.116020
CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes
Abstract
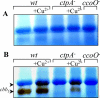
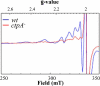
The ctpA (ccoI) gene product, a putative inner membrane copper-translocating P1B-type ATPase present in many bacteria, has been shown to be involved only in the cbb(3) assembly in Rhodobacter capsulatus and Bradyrhizobium japonicum. ctpA was disrupted in Rubrivivax gelatinosus, and the mutants showed a drastic decrease in both cbb(3) and caa(3) oxidase activities. Inactivation of ctpA results also in a decrease in the amount of the nitrous oxide reductase, NosZ. This pleiotropic phenotype could be partially rescued by excess copper in the medium, indicating that CtpA is likely a copper transporter that supplies copper-requiring proteins in the membrane with this metal. Although CtpA shares significant sequence homologies with the homeostasis copper efflux P1B-type ATPases including the bacterial CopA and the human ATP7A and ATP7B, disruption of ctpA did not result in any sensitivity to excess copper. This indicates that the CtpA is not crucial for copper tolerance but is involved in the assembly of membrane and periplasmic copper enzymes in this bacterium. The potential roles of CtpA in bacteria in comparison with CopA are discussed.
Figures


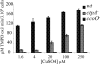



References
-
- Puig S., Thiele D. J. (2002) Curr. Opin. Chem. Biol. 6, 171–180 - PubMed
-
- Zumft W. G., Kroneck P. M. (2007) Adv. Microb. Physiol. 52, 107–227 - PubMed
-
- Ostermeier C., Iwata S., Michel H. (1996) Curr. Opin. Struct. Biol. 6, 460–466 - PubMed
-
- Pitcher R. S., Watmough N. J. (2004) Biochim. Biophys. Acta 1655, 388–399 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

