Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice
- PMID: 20364083
- PMCID: PMC2860901
- DOI: 10.1172/JCI38331
Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice
Abstract
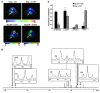
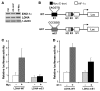
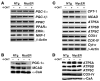
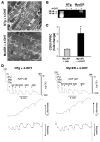
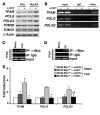
In the adult heart, regulation of fatty acid oxidation and mitochondrial genes is controlled by the PPARgamma coactivator-1 (PGC-1) family of transcriptional coactivators. However, in response to pathological stressors such as hemodynamic load or ischemia, cardiac myocytes downregulate PGC-1 activity and fatty acid oxidation genes in preference for glucose metabolism pathways. Interestingly, despite the reduced PGC-1 activity, these pathological stressors are associated with mitochondrial biogenesis, at least initially. The transcription factors that regulate these changes in the setting of reduced PGC-1 are unknown, but Myc can regulate glucose metabolism and mitochondrial biogenesis during cell proliferation and tumorigenesis in cancer cells. Here we have demonstrated that Myc activation in the myocardium of adult mice increases glucose uptake and utilization, downregulates fatty acid oxidation by reducing PGC-1alpha levels, and induces mitochondrial biogenesis. Inactivation of Myc in the adult myocardium attenuated hypertrophic growth and decreased the expression of glycolytic and mitochondrial biogenesis genes in response to hemodynamic load. Surprisingly, the Myc-orchestrated metabolic alterations were associated with preserved cardiac function and improved recovery from ischemia. Our data suggest that Myc directly regulates glucose metabolism and mitochondrial biogenesis in cardiac myocytes and is an important regulator of energy metabolism in the heart in response to pathologic stress.
Figures
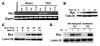
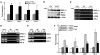

References
-
- Lopaschuk GD, Spafford MA. Energy substrate utilization by isolated working hearts from newborn rabbits. Am J Physiol. 1990;258(5 Pt 2):H1274–H1280. - PubMed
-
- Spitkovsky D, et al. Activity of complex III of the mitochondrial electron transport chain is essential for early heart muscle cell differentiation. FASEB J. 2004;18(11):1300–1302. - PubMed
-
- Schwartz K, Boheler KR, de la Bastie D, Lompre AM, Mercadier JJ. Switches in cardiac muscle gene expression as a result of pressure and volume overload. Am J Physiol. 1992;262(3 pt 2):R364–R369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

