Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
- PMID: 20364130
- PMCID: PMC3280089
- DOI: 10.1038/nsmb.1800
Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
Abstract
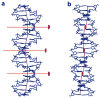
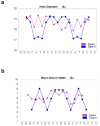
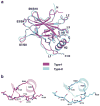
p53 binds as a tetramer to DNA targets consisting of two decameric half-sites separated by a variable spacer. Here we present high-resolution crystal structures of complexes between p53 core-domain tetramers and DNA targets consisting of contiguous half-sites. In contrast to previously reported p53-DNA complexes that show standard Watson-Crick base pairs, the newly reported structures show noncanonical Hoogsteen base-pairing geometry at the central A-T doublet of each half-site. Structural and computational analyses show that the Hoogsteen geometry distinctly modulates the B-DNA helix in terms of local shape and electrostatic potential, which, together with the contiguous DNA configuration, results in enhanced protein-DNA and protein-protein interactions compared to noncontiguous half-sites. Our results suggest a mechanism relating spacer length to protein-DNA binding affinity. Our findings also expand the current understanding of protein-DNA recognition and establish the structural and chemical properties of Hoogsteen base pairs as the basis for a novel mode of sequence readout.
Figures
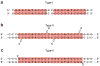
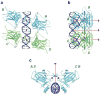
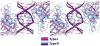
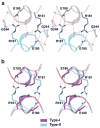

Comment in
-
Four p(53)s in a pod.Nat Struct Mol Biol. 2010 Apr;17(4):390-1. doi: 10.1038/nsmb0410-390. Nat Struct Mol Biol. 2010. PMID: 20368720 No abstract available.
References
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. - PubMed
-
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. - PubMed
-
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–42. - PubMed
-
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–61. - PubMed
-
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature Gen. 1992;1:45–49. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

