The exon junction complex component Magoh controls brain size by regulating neural stem cell division
- PMID: 20364144
- PMCID: PMC2860667
- DOI: 10.1038/nn.2527
The exon junction complex component Magoh controls brain size by regulating neural stem cell division
Abstract
Brain structure and size require precise division of neural stem cells (NSCs), which self-renew and generate intermediate neural progenitors (INPs) and neurons. The factors that regulate NSCs remain poorly understood, and mechanistic explanations of how aberrant NSC division causes the reduced brain size seen in microcephaly are lacking. Here we show that Magoh, a component of the exon junction complex (EJC) that binds RNA, controls mouse cerebral cortical size by regulating NSC division. Magoh haploinsufficiency causes microcephaly because of INP depletion and neuronal apoptosis. Defective mitosis underlies these phenotypes, as depletion of EJC components disrupts mitotic spindle orientation and integrity, chromosome number and genomic stability. In utero rescue experiments showed that a key function of Magoh is to control levels of the microcephaly-associated protein Lis1 during neurogenesis. Our results uncover requirements for the EJC in brain development, NSC maintenance and mitosis, thereby implicating this complex in the pathogenesis of microcephaly.
Figures
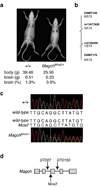
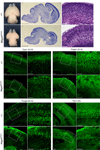
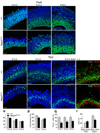
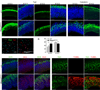
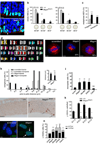
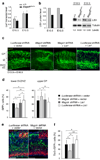
Comment in
-
Regulating brain size.Nat Neurosci. 2010 May;13(5):531. doi: 10.1038/nn0510-531. Nat Neurosci. 2010. PMID: 20421899 No abstract available.
References
-
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. - PubMed
-
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;Vol. 30:24–32. - PubMed
-
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. - PubMed
-
- Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

