Changes in the accessibility of the HIV-1 Integrase C-terminus in the presence of cellular proteins
- PMID: 20367881
- PMCID: PMC2859751
- DOI: 10.1186/1742-4690-7-27
Changes in the accessibility of the HIV-1 Integrase C-terminus in the presence of cellular proteins
Abstract
Background: Following entry, uncoating, and reverse transcription, a number of cellular proteins become associated with the human immunodeficiency virus type 1 (HIV-1) pre-integration complex (PIC). With the goal of obtaining reagents for the analysis of the HIV-1 PIC composition and localisation, we have constructed functional integrase (IN) and matrix (MA) proteins that can be biotinylated during virus production and captured using streptavidin-coated beads.
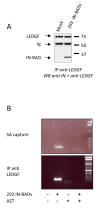
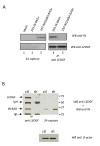
Results: Although the labelled C-terminus allows for the sensitive detection of virion-associated IN, it becomes inaccessible in the presence of cellular proteins. This masking is not dependent on the nature of the tag and does not occur with the tagged MA. It was not observed either with an IN mutant unable to interact with LEDGF/p75, or when LEDGF/p75 was depleted from cells.
Conclusion: Our observation suggests that a structural rearrangement or oligomerization of the IN protein occurs during the early steps of infection and that this process is related to the presence of LEDGF/p75.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

