Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme
- PMID: 20368332
- PMCID: PMC2888440
- DOI: 10.1074/jbc.M110.102921
Crystal structure of the human ubiquitin-activating enzyme 5 (UBA5) bound to ATP: mechanistic insights into a minimalistic E1 enzyme
Abstract
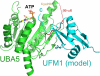
E1 ubiquitin-activating enzymes (UBAs) are large multidomain proteins that catalyze formation of a thioester bond between the terminal carboxylate of a ubiquitin or ubiquitin-like modifier (UBL) and a conserved cysteine in an E2 protein, producing reactive ubiquityl units for subsequent ligation to substrate lysines. Two important E1 reaction intermediates have been identified: a ubiquityl-adenylate phosphoester and a ubiquityl-enzyme thioester. However, the mechanism of thioester bond formation and its subsequent transfer to an E2 enzyme remains poorly understood. We have determined the crystal structure of the human UFM1 (ubiquitin-fold modifier 1) E1-activating enzyme UBA5, bound to ATP, revealing a structure that shares similarities with both large canonical E1 enzymes and smaller ancestral E1-like enzymes. In contrast to other E1 active site cysteines, which are in a variably sized domain that is separate and flexible relative to the adenylation domain, the catalytic cysteine of UBA5 (Cys(250)) is part of the adenylation domain in an alpha-helical motif. The novel position of the UBA5 catalytic cysteine and conformational changes associated with ATP binding provides insight into the possible mechanisms through which the ubiquityl-enzyme thioester is formed. These studies reveal structural features that further our understanding of the UBA5 enzyme reaction mechanism and provide insight into the evolution of ubiquitin activation.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

