Evolutionary dynamics of Clostridium difficile over short and long time scales
- PMID: 20368420
- PMCID: PMC2867753
- DOI: 10.1073/pnas.0914322107
Evolutionary dynamics of Clostridium difficile over short and long time scales
Abstract
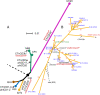
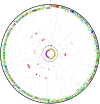
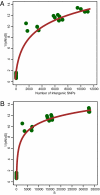
Clostridium difficile has rapidly emerged as the leading cause of antibiotic-associated diarrheal disease, with the transcontinental spread of various PCR ribotypes, including 001, 017, 027 and 078. However, the genetic basis for the emergence of C. difficile as a human pathogen is unclear. Whole genome sequencing was used to analyze genetic variation and virulence of a diverse collection of thirty C. difficile isolates, to determine both macro and microevolution of the species. Horizontal gene transfer and large-scale recombination of core genes has shaped the C. difficile genome over both short and long time scales. Phylogenetic analysis demonstrates C. difficile is a genetically diverse species, which has evolved within the last 1.1-85 million years. By contrast, the disease-causing isolates have arisen from multiple lineages, suggesting that virulence evolved independently in the highly epidemic lineages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298:531–534. - PubMed
-
- Bartlett JG. Narrative review: The new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med. 2006;145:758–764. - PubMed
-
- Cheknis AK, et al. Distribution of Clostridium difficile strains from a North American, European and Australian trial of treatment for C. difficile infections: 2005-2007. Anaerobe. 2009;15:230–233. - PubMed
-
- Brazier JS, Patel B, Pearson A. Distribution of Clostridium difficile PCR ribotype 027 in British hospitals. Eur Surveill. 2007;12 pii=3182. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

