Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing
- PMID: 20368428
- PMCID: PMC2867740
- DOI: 10.1073/pnas.1003055107
Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing
Abstract
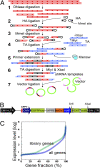
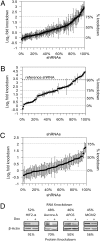
As a general strategy for function-based gene identification, an shRNA library containing approximately 150 shRNAs per gene was enzymatically generated from normalized (reduced-redundance) human cDNA. The library was constructed in an inducible lentiviral vector, enabling propagation of growth-inhibiting shRNAs and controlled activity measurements. RNAi activities were measured for 101 shRNA clones representing 100 human genes and for 201 shRNAs derived from a firefly luciferase gene. Structure-activity analysis of these two datasets yielded a set of structural criteria for shRNA efficacy, increasing the frequencies of active shRNAs up to 5-fold relative to random sampling. The same library was used to select shRNAs that inhibit breast carcinoma cell growth by targeting potential oncogenes. Genes targeted by the selected shRNAs were enriched for 10 pathways, 9 of which have been previously associated with various cancers, cell cycle progression, or apoptosis. One hundred nineteen genes, enriched through this selection and represented by two to six shRNAs each, were identified as potential cancer drug targets. Short interfering RNAs against 19 of 22 tested genes in this group inhibited cell growth, validating the efficiency of this strategy for high-throughput target gene identification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
-
- Pei Y, Tuschl T. On the art of identifying effective and specific siRNAs. Nat Methods. 2006;3:670–676. - PubMed
-
- Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. - PubMed
-
- Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

