Contrasting genetic paths to morphological and physiological evolution
- PMID: 20368429
- PMCID: PMC2867737
- DOI: 10.1073/pnas.0910339107
Contrasting genetic paths to morphological and physiological evolution
Abstract
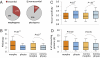
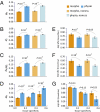
The relative importance of protein function change and gene expression change in phenotypic evolution is a contentious, yet central topic in evolutionary biology. Analyzing 5,199 mouse genes with recorded mutant phenotypes, we find that genes exclusively affecting morphological traits when mutated (dubbed "morphogenes") are grossly enriched with transcriptional regulators, whereas those exclusively affecting physiological traits (dubbed "physiogenes") are enriched with channels, transporters, receptors, and enzymes. Compared to physiogenes, morphogenes are more likely to be essential and pleiotropic and less likely to be tissue specific. Morphogenes evolve faster in expression profile, but slower in protein sequence and gene gain/loss than physiogenes. Thus, morphological and physiological changes have a differential molecular basis; separating them helps discern the genetic mechanisms of phenotypic evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
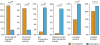
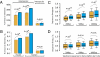
References
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Glazko G, Veeramachaneni V, Nei M, Makałowski W. Eighty percent of proteins are different between humans and chimpanzees. Gene. 2005;346:215–219. - PubMed
-
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. - PubMed
-
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

