Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots
- PMID: 20368455
- PMCID: PMC2867735
- DOI: 10.1073/pnas.0908269107
Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots
Abstract
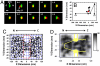
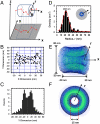
The translocation of large macromolecules through the nuclear pore complex (NPC) of eukaryotic cells is hindered by the phenylalanine-glycine (FG) nucleoporin (Nup) barrier unless molecules are chaperoned by transport receptors. The precise mechanism of facilitated translocation remains unclear due to the challenges of measuring the series of transient interactions between a transport receptor and the FG-Nups. This study developed single-point edge-excitation subdiffraction microscopy to obtain a three-dimensional density map of the transient interactions with a spatiotemporal resolution of 9 nm and 400 mus. Three unique features were observed under real-time trafficking conditions that have escaped detection by conventional electron microscopy: (i) the spatial density of interaction sites between Importin beta1 (Imp beta1, a major transport receptor) and the FG-Nups gradually increases from both sides of the NPC and is highest in the central pore region; (ii) cargo-free or cargo-bound Imp beta1 rarely occupies an axial channel with a diameter of approximately 10-20 nm at its narrowest point through the NPC; and (iii) the pathway of facilitated translocation through the NPC depends more on the interaction sites of the FG-Nups than on the NPC architecture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Weis K. Regulating access to the genome: Nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. - PubMed
-
- Fahrenkrog B, Aebi U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. - PubMed
-
- Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

