Metamorphic proteins mediate evolutionary transitions of structure
- PMID: 20368465
- PMCID: PMC2867682
- DOI: 10.1073/pnas.0912616107
Metamorphic proteins mediate evolutionary transitions of structure
Abstract
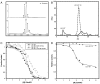
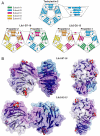
The primary sequence of proteins usually dictates a single tertiary and quaternary structure. However, certain proteins undergo reversible backbone rearrangements. Such metamorphic proteins provide a means of facilitating the evolution of new folds and architectures. However, because natural folds emerged at the early stages of evolution, the potential role of metamorphic intermediates in mediating evolutionary transitions of structure remains largely unexplored. We evolved a set of new proteins based on approximately 100 amino acid fragments derived from tachylectin-2--a monomeric, 236 amino acids, five-bladed beta-propeller. Their structures reveal a unique pentameric assembly and novel beta-propeller structures. Although identical in sequence, the oligomeric subunits adopt two, or even three, different structures that together enable the pentameric assembly of two propellers connected via a small linker. Most of the subunits adopt a wild-type-like structure within individual five-bladed propellers. However, the bridging subunits exhibit domain swaps and asymmetric strand exchanges that allow them to complete the two propellers and connect them. Thus, the modular and metamorphic nature of these subunits enabled dramatic changes in tertiary and quaternary structure, while maintaining the lectin function. These oligomers therefore comprise putative intermediates via which beta-propellers can evolve from smaller elements. Our data also suggest that the ability of one sequence to equilibrate between different structures can be evolutionary optimized, thus facilitating the emergence of new structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Reconstruction of functional beta-propeller lectins via homo-oligomeric assembly of shorter fragments.J Mol Biol. 2007 Jan 5;365(1):10-7. doi: 10.1016/j.jmb.2006.09.055. Epub 2006 Sep 27. J Mol Biol. 2007. PMID: 17054983
-
Functional β-propeller lectins by tandem duplications of repetitive units.Protein Eng Des Sel. 2011 Jan;24(1-2):185-95. doi: 10.1093/protein/gzq053. Epub 2010 Aug 16. Protein Eng Des Sel. 2011. PMID: 20713410
-
De Novo Evolutionary Emergence of a Symmetrical Protein Is Shaped by Folding Constraints.Cell. 2016 Jan 28;164(3):476-86. doi: 10.1016/j.cell.2015.12.024. Epub 2016 Jan 21. Cell. 2016. PMID: 26806127 Free PMC article.
-
Insights into the quaternary association of proteins through structure graphs: a case study of lectins.Biochem J. 2005 Oct 1;391(Pt 1):1-15. doi: 10.1042/BJ20050434. Biochem J. 2005. PMID: 16173917 Free PMC article. Review.
-
Fold change in evolution of protein structures.J Struct Biol. 2001 May-Jun;134(2-3):167-85. doi: 10.1006/jsbi.2001.4335. J Struct Biol. 2001. PMID: 11551177 Review.
Cited by
-
Probing pathways of adaptation with continuous evolution.Curr Opin Syst Biol. 2019 Apr;14:18-24. doi: 10.1016/j.coisb.2019.02.002. Epub 2019 Feb 13. Curr Opin Syst Biol. 2019. PMID: 31608311 Free PMC article. No abstract available.
-
Development and applications of artificial symmetrical proteins.Comput Struct Biotechnol J. 2020 Nov 27;18:3959-3968. doi: 10.1016/j.csbj.2020.10.040. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33335692 Free PMC article. Review.
-
Why reinvent the wheel? Building new proteins based on ready-made parts.Protein Sci. 2016 Jul;25(7):1179-87. doi: 10.1002/pro.2892. Epub 2016 Feb 22. Protein Sci. 2016. PMID: 26821641 Free PMC article. Review.
-
Fragile protein folds: Sequence and environmental factors affecting the equilibrium of two interconverting, stably folded protein conformations.Magn Reson (Gott). 2021;2(1):63-76. doi: 10.5194/mr-2-63-2021. Epub 2021 Mar 10. Magn Reson (Gott). 2021. PMID: 35603043 Free PMC article.
-
Extant fold-switching proteins are widespread.Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):5968-5973. doi: 10.1073/pnas.1800168115. Epub 2018 May 21. Proc Natl Acad Sci U S A. 2018. PMID: 29784778 Free PMC article.
References
-
- Maynard-Smith J. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. - PubMed
-
- Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. - PubMed
-
- Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet. 2006;38:168–174. - PubMed
-
- Meier S, Ozbek S. A biological cosmos of parallel universes: does protein structural plasticity facilitate evolution? Bioessays. 2007;29:1095–1104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

