53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination
- PMID: 20368578
- PMCID: PMC2856023
- DOI: 10.1084/jem.20100244
53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination
Abstract
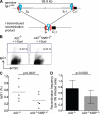
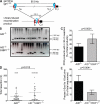
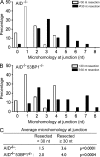
Class switch recombination (CSR) diversifies antibodies by joining highly repetitive DNA elements, which are separated by 60-200 kbp. CSR is initiated by activation-induced cytidine deaminase, an enzyme that produces multiple DNA double-strand breaks (DSBs) in switch regions. Switch regions are joined by a mechanism that requires an intact DNA damage response and classical or alternative nonhomologous end joining (A-NHEJ). Among the DNA damage response factors, 53BP1 has the most profound effect on CSR. We explore the role of 53BP1 in intrachromosomal DNA repair using I-SceI to introduce paired DSBs in the IgH locus. We find that the absence of 53BP1 results in an ataxia telangiectasia mutated-dependent increase in DNA end resection and that resected DNA is preferentially repaired by microhomology-mediated A-NHEJ. We propose that 53BP1 favors long-range CSR in part by protecting DNA ends against resection, which prevents A-NHEJ-dependent short-range rejoining of intra-switch region DSBs.
Figures



References
-
- Boboila C., Jankovic M., Yan C.T., Wang J.H., Wesemann D.R., Zhang T., Fazeli A., Feldman L., Nussenzweig A., Nussenzweig M., Alt F.W. 2010a. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA. 107:3034–3039 - PMC - PubMed
-
- Catalan N., Selz F., Imai K., Revy P., Fischer A., Durandy A. 2003. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 171:2504–2509 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

