CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response
- PMID: 20368616
- PMCID: PMC2854379
- DOI: 10.1083/jcb.200912163
CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response
Abstract
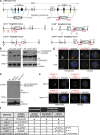
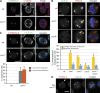
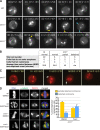
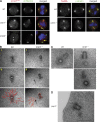
The centrosomal protein, CDK5RAP2, is mutated in primary microcephaly, a neurodevelopmental disorder characterized by reduced brain size. The Drosophila melanogaster homologue of CDK5RAP2, centrosomin (Cnn), maintains the pericentriolar matrix (PCM) around centrioles during mitosis. In this study, we demonstrate a similar role for CDK5RAP2 in vertebrate cells. By disrupting two evolutionarily conserved domains of CDK5RAP2, CNN1 and CNN2, in the avian B cell line DT40, we find that both domains are essential for linking centrosomes to mitotic spindle poles. Although structurally intact, centrosomes lacking the CNN1 domain fail to recruit specific PCM components that mediate attachment to spindle poles. Furthermore, we show that the CNN1 domain enforces cohesion between parental centrioles during interphase and promotes efficient DNA damage-induced G2 cell cycle arrest. Because mitotic spindle positioning, asymmetric centrosome inheritance, and DNA damage signaling have all been implicated in cell fate determination during neurogenesis, our findings provide novel insight into how impaired CDK5RAP2 function could cause premature depletion of neural stem cells and thereby microcephaly.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

