Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes
- PMID: 20368972
- PMCID: PMC2848560
- DOI: 10.1371/journal.ppat.1000848
Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes
Abstract
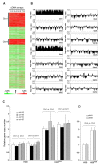
Cryptococcus neoformans is a haploid environmental organism and the major cause of fungal meningoencephalitis in AIDS patients. Fluconazole (FLC), a triazole, is widely used for the maintenance therapy of cryptococcosis. Heteroresistance to FLC, an adaptive mode of azole resistance, was associated with FLC therapy failure cases but the mechanism underlying the resistance was unknown. We used comparative genome hybridization and quantitative real-time PCR in order to show that C. neoformans adapts to high concentrations of FLC by duplication of multiple chromosomes. Formation of disomic chromosomes in response to FLC stress was observed in both serotype A and D strains. Strains that adapted to FLC concentrations higher than their minimal inhibitory concentration (MIC) contained disomies of chromosome 1 and stepwise exposure to even higher drug concentrations induced additional duplications of several other specific chromosomes. The number of disomic chromosomes in each resistant strain directly correlated with the concentration of FLC tolerated by each strain. Upon removal of the drug pressure, strains that had adapted to high concentrations of FLC returned to their original level of susceptibility by initially losing the extra copy of chromosome 1 followed by loss of the extra copies of the remaining disomic chromosomes. The duplication of chromosome 1 was closely associated with two of its resident genes: ERG11, the target of FLC and AFR1, the major transporter of azoles in C. neoformans. This adaptive mechanism in C. neoformans may play an important role in FLC therapy failure of cryptococcosis leading to relapse during azole maintenance therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

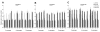


References
-
- Kwon-Chung KJ, Bennett JE. Philadelphia: Lea & Febiger; 1992. Medical Mycology.866
-
- Perfect JR, Casadevall A. Cryptococcosis. Infect Dis Clin North Am. 2002;16:837–874. - PubMed
-
- Zonios DI, Bennett JE. Update on azole antifungals. Semin Respir Crit Care Med. 2008;29:198–210. - PubMed
-
- Perfect JR, Cox GM. Drug resistance in Cryptococcus neoformans. Drug Resist Updat. 1999;2:259–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

