Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis
- PMID: 20368982
- PMCID: PMC2848570
- DOI: 10.1371/journal.pone.0009875
Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis
Abstract
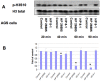
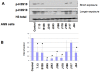
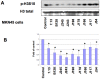
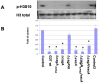
Histone modifications are critical in regulating gene expression, cell cycle, cell proliferation, and development. Relatively few studies have investigated whether Helicobacter pylori, the major cause of human gastric diseases, affects histone modification. We therefore investigated the effects of H. pylori infection on histone modifications in a global and promoter-specific manner in gastric epithelial cells. Infection of gastric epithelial cells by wild-type H. pylori induced time- and dose-dependent dephosphorylation of histone H3 at serine 10 (H3 Ser10) and decreased acetylation of H3 lysine 23, but had no effects on seven other specific modifications. Different cag pathogenicity island (PAI)-containing-clinical isolates showed similar abilities to induce H3 Ser10 dephosphorylation. Mutation of cagA, vacA, nonphosphorylateable CagA mutant cagA(EPISA), or disruption of the flagella showed no effects, while deletion of the entire cagPAI restored the H3 Ser10 phosphorylation to control levels. Analysis of 27 cagPAI mutants indicated that the genes that caused H3 Ser10 dephosphorylation were similar to those that were previously found to induce interleukin-8, irrespective of CagA translocation. This effect was independent of ERK or p38 pathways and type I interferon signaling. Additionally, c-Jun and hsp70 gene expression was associated with this histone modification. These results demonstrate that H. pylori alters histone modification and host response via a cagA-, vacA-independent, but cagPAI-dependent mechanisms, which contribute to its persistent infection and pathogenesis.
Conflict of interest statement
Figures








Similar articles
-
Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation.Epigenetics. 2009 Nov 16;4(8):577-86. doi: 10.4161/epi.4.8.10217. Epigenetics. 2009. PMID: 20081355
-
CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation.PLoS Pathog. 2015 Feb 6;11(2):e1004663. doi: 10.1371/journal.ppat.1004663. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25658601 Free PMC article.
-
Helicobacter pylori cag pathogenicity island-positive strains induce syndecan-4 expression in gastric epithelial cells.FEMS Immunol Med Microbiol. 2009 Aug;56(3):223-32. doi: 10.1111/j.1574-695X.2009.00569.x. Epub 2009 May 16. FEMS Immunol Med Microbiol. 2009. PMID: 19519784
-
[High prevalence of VacA, CagA protein expression and presence of cag pathogenicity island in Japanese Helicobacter pylori isolates].Nihon Rinsho. 1999 Jan;57(1):17-22. Nihon Rinsho. 1999. PMID: 10036931 Review. Japanese.
-
Pathogenic mechanisms of the oncoprotein CagA in H. pylori-induced gastric cancer (Review).Oncol Rep. 2016 Dec;36(6):3087-3094. doi: 10.3892/or.2016.5145. Epub 2016 Oct 4. Oncol Rep. 2016. PMID: 27748858 Review.
Cited by
-
Epigenetic regulation in bacterial infections: targeting histone deacetylases.Crit Rev Microbiol. 2018 May;44(3):336-350. doi: 10.1080/1040841X.2017.1373063. Epub 2017 Oct 3. Crit Rev Microbiol. 2018. PMID: 28971711 Free PMC article. Review.
-
Helicobacter pylori infection-induced H3Ser10 phosphorylation in stepwise gastric carcinogenesis and its clinical implications.Helicobacter. 2018 Jun;23(3):e12486. doi: 10.1111/hel.12486. Epub 2018 Apr 15. Helicobacter. 2018. PMID: 29656498 Free PMC article.
-
The Microbiological Memory, an Epigenetic Regulator Governing the Balance Between Good Health and Metabolic Disorders.Front Microbiol. 2018 Jun 26;9:1379. doi: 10.3389/fmicb.2018.01379. eCollection 2018. Front Microbiol. 2018. PMID: 29997595 Free PMC article.
-
Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases.Clin Epigenetics. 2015 Oct 16;7:112. doi: 10.1186/s13148-015-0144-7. eCollection 2015. Clin Epigenetics. 2015. PMID: 26478753 Free PMC article. Review.
-
Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation.Carcinogenesis. 2013 Sep;34(9):1955-67. doi: 10.1093/carcin/bgt212. Epub 2013 Jun 7. Carcinogenesis. 2013. PMID: 23749751 Free PMC article. Review.
References
-
- Peek RM, Jr,, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208(2):233–248. - PubMed
-
- Ernst PB, Peura DA, Crowe SE. The translation of Helicobacter pylori basic research to patient care. Gastroenterology. 2006;130(1):188–206; quiz; 212-3. - PubMed
-
- Lu H, Yamaoka Y, Graham DY. Helicobacter pylori virulence factors: Facts and fantasies. Curr Opin Gastroenterol. 2005;21(6):653–659. - PubMed
-
- Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301(5636):1099–1102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

