Differential transcript expression between the microfilariae of the filarial nematodes, Brugia malayi and B. pahangi
- PMID: 20370932
- PMCID: PMC2874553
- DOI: 10.1186/1471-2164-11-225
Differential transcript expression between the microfilariae of the filarial nematodes, Brugia malayi and B. pahangi
Abstract
Background: Brugia malayi and B. pahangi are two closely related nematodes that cause filariasis in humans and animals. However, B. pahangi microfilariae are able to develop in and be transmitted by the mosquito, Armigeres subalbatus, whereas most B. malayi are rapidly melanized and destroyed within the mosquito hemocoel. A cross-species microarray analysis employing the B. malayi V2 array was carried out to determine the transcriptional differences between B. malayi and B. pahangi microfilariae with similar age distribution.
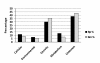
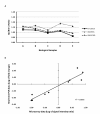
Results: Following microarray data analysis, a list of preferentially expressed genes in both microfilariae species was generated with a false discovery rate estimate of 5% and a signal intensity ratio of 2 or higher in either species. A total of 308 probes were preferentially expressed in both species with 149 probes, representing 123 genes, in B. pahangi microfilariae and 159 probes, representing 107 genes, in B. malayi microfilariae. In B. pahangi, there were 76 (62%) up-regulated transcripts that coded for known proteins that mapped into the KEGG pathway compared to 61 (57%) transcripts in B. malayi microfilariae. The remaining 47 (38%) transcripts in B. pahangi and 46 (43%) transcripts in B. malayi microfilariae were comprised almost entirely of hypothetical genes of unknown function. Twenty-seven of the transcripts in B. pahangi microfilariae coded for proteins that associate with the secretory pathway compared to thirty-nine in B. malayi microfilariae. The data obtained from real-time PCR analysis of ten genes selected from the microarray list of preferentially expressed genes showed good concordance with the microarray data, indicating that the microarray data were reproducible.
Conclusion: In this study, we identified gene transcripts that were preferentially expressed in the microfilariae of B. pahangi and B. malayi, some of which coded for known immunomodulatory proteins. These comparative transcriptome data will be of interest to researchers keen on understanding the inherent differences, at the molecular level, between B. malayi and B. pahangi microfilariae especially because these microfilariae are capable of surviving in the same vertebrate host but elicit different immune response outcomes in the mosquito, Ar. subalbatus.
Figures

Similar articles
-
Mosquito transcriptome profiles and filarial worm susceptibility in Armigeres subalbatus.PLoS Negl Trop Dis. 2010 Apr 20;4(4):e666. doi: 10.1371/journal.pntd.0000666. PLoS Negl Trop Dis. 2010. PMID: 20421927 Free PMC article.
-
Mosquito transcriptome changes and filarial worm resistance in Armigeres subalbatus.BMC Genomics. 2007 Dec 18;8:463. doi: 10.1186/1471-2164-8-463. BMC Genomics. 2007. PMID: 18088420 Free PMC article.
-
Response of Armigeres subalbatus (Diptera: Culicidae) to intraperitoneally isolated Brugia spp. microfilariae.J Med Entomol. 2007 Mar;44(2):295-8. doi: 10.1603/0022-2585(2007)44[295:roasdc]2.0.co;2. J Med Entomol. 2007. PMID: 17427699
-
Recent advances in the application of molecular biology in filariasis.Southeast Asian J Trop Med Public Health. 1993;24 Suppl 2:55-63. Southeast Asian J Trop Med Public Health. 1993. PMID: 7973949 Review.
-
Examining the role of macrolides and host immunity in combatting filarial parasites.Parasit Vectors. 2017 Apr 14;10(1):182. doi: 10.1186/s13071-017-2116-6. Parasit Vectors. 2017. PMID: 28410595 Free PMC article. Review.
Cited by
-
The Symbiotic Bacteria-Xenorhabdus nematophila All and Photorhabdus luminescens H06 Strongly Affected the Phenoloxidase Activation of Nipa Palm Hispid, Octodonta nipae (Coleoptera: Chrysomelidae) Larvae.Pathogens. 2023 Mar 23;12(4):506. doi: 10.3390/pathogens12040506. Pathogens. 2023. PMID: 37111392 Free PMC article.
-
Extracellular vesicles secreted by Brugia malayi microfilariae modulate the melanization pathway in the mosquito host.Sci Rep. 2023 May 31;13(1):8778. doi: 10.1038/s41598-023-35940-9. Sci Rep. 2023. PMID: 37258694 Free PMC article.
-
The role of 'omics' in the quest to eliminate human filariasis.PLoS Negl Trop Dis. 2017 Apr 20;11(4):e0005464. doi: 10.1371/journal.pntd.0005464. eCollection 2017 Apr. PLoS Negl Trop Dis. 2017. PMID: 28426656 Free PMC article. Review. No abstract available.
-
Transcriptome-wide analysis of filarial extract-primed human monocytes reveal changes in LPS-induced PTX3 expression levels.Sci Rep. 2019 Feb 22;9(1):2562. doi: 10.1038/s41598-019-38985-x. Sci Rep. 2019. PMID: 30796272 Free PMC article.
-
Parallel Evolution of C-Type Lectin Domain Gene Family Sizes in Insect-Vectored Nematodes.Front Plant Sci. 2022 Apr 25;13:856826. doi: 10.3389/fpls.2022.856826. eCollection 2022. Front Plant Sci. 2022. PMID: 35557736 Free PMC article.
References
-
- Mak JW. Epidemiology of lymphatic filariasis. Ciba Found Symp. 1987;127:5–14. - PubMed
-
- McReynolds LA, Poole C, Hong Y, Williams SA, Partono F, Bradley J. Recent advances in the application of molecular biology in filariasis. Southeast Asian J Trop Med Public Health. 1993;24(Suppl 2):55–63. - PubMed
-
- Xie H, Bain O, Williams SA. Molecular phylogenetic studies on Brugia filariae using Hha I repeat sequences. Parasite. 1994;1(3):255–260. - PubMed
-
- Nayar JK, Knight JW, Vickery AC. Susceptibility of Anopheles quadrimaculatus (Diptera: Culicidae) to subperiodic Brugia malayi and Brugia pahangi (Nematoda: Filarioidea) adapted to nude mice and jirds. J Med Entomol. 1990;27(3):409–411. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

