Mechanical unfolding of an ankyrin repeat protein
- PMID: 20371329
- PMCID: PMC2849098
- DOI: 10.1016/j.bpj.2009.12.4287
Mechanical unfolding of an ankyrin repeat protein
Abstract
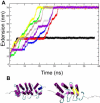
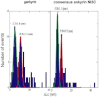
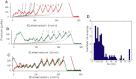
Ankryin repeat proteins comprise tandem arrays of a 33-residue, predominantly alpha-helical motif that stacks roughly linearly to produce elongated and superhelical structures. They function as scaffolds mediating a diverse range of protein-protein interactions, and some have been proposed to play a role in mechanical signal transduction processes in the cell. Here we use atomic force microscopy and molecular-dynamics simulations to investigate the natural 7-ankyrin repeat protein gankyrin. We find that gankyrin unfolds under force via multiple distinct pathways. The reactions do not proceed in a cooperative manner, nor do they always involve fully stepwise unfolding of one repeat at a time. The peeling away of half an ankyrin repeat, or one or more ankyrin repeats, occurs at low forces; however, intermediate species are formed that are resistant to high forces, and the simulations indicate that in some instances they are stabilized by nonnative interactions. The unfolding of individual ankyrin repeats generates a refolding force, a feature that may be more easily detected in these proteins than in globular proteins because the refolding of a repeat involves a short contraction distance and incurs a low entropic cost. We discuss the origins of the differences between the force- and chemical-induced unfolding pathways of ankyrin repeat proteins, as well as the differences between the mechanics of natural occurring ankyrin repeat proteins and those of designed consensus ankyin repeat and globular proteins.
Copyright (c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Spectrin domains lose cooperativity in forced unfolding.Biophys J. 2007 Jan 15;92(2):571-7. doi: 10.1529/biophysj.106.093690. Epub 2006 Nov 3. Biophys J. 2007. PMID: 17085494 Free PMC article.
-
Reconstruction of mechanical unfolding and refolding pathways of proteins with atomic force spectroscopy and computer simulations.Methods. 2022 Jan;197:39-53. doi: 10.1016/j.ymeth.2021.05.012. Epub 2021 May 18. Methods. 2022. PMID: 34020035
-
Effects of ligand binding on the mechanical properties of ankyrin repeat protein gankyrin.PLoS Comput Biol. 2013;9(1):e1002864. doi: 10.1371/journal.pcbi.1002864. Epub 2013 Jan 17. PLoS Comput Biol. 2013. PMID: 23341763 Free PMC article.
-
Protein unfolding and refolding under force: methodologies for nanomechanics.Chemphyschem. 2005 Jan;6(1):29-34. doi: 10.1002/cphc.200400343. Chemphyschem. 2005. PMID: 15688640 Review.
-
Folding and Stability of Ankyrin Repeats Control Biological Protein Function.Biomolecules. 2021 Jun 5;11(6):840. doi: 10.3390/biom11060840. Biomolecules. 2021. PMID: 34198779 Free PMC article. Review.
Cited by
-
Prying open single GroES ring complexes by force reveals cooperativity across domains.Biophys J. 2012 Apr 18;102(8):1961-8. doi: 10.1016/j.bpj.2012.03.046. Biophys J. 2012. PMID: 22768953 Free PMC article.
-
Resolving the fine structure in the energy landscapes of repeat proteins.QRB Discov. 2022 Jun 10;3:e7. doi: 10.1017/qrd.2022.4. eCollection 2022. QRB Discov. 2022. PMID: 37529289 Free PMC article.
-
Distinct roles of TRP channels in auditory transduction and amplification in Drosophila.Neuron. 2013 Jan 9;77(1):115-28. doi: 10.1016/j.neuron.2012.11.030. Neuron. 2013. PMID: 23312520 Free PMC article.
-
Mechanical anisotropy of ankyrin repeats.Biophys J. 2012 Mar 7;102(5):1118-26. doi: 10.1016/j.bpj.2012.01.046. Epub 2012 Mar 6. Biophys J. 2012. PMID: 22404934 Free PMC article.
-
Full reconstruction of a vectorial protein folding pathway by atomic force microscopy and molecular dynamics simulations.J Biol Chem. 2010 Dec 3;285(49):38167-72. doi: 10.1074/jbc.M110.179697. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870713 Free PMC article.
References
-
- Binz H.K., Stumpp M.T., Plückthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003;332:489–503. - PubMed
-
- Bornschlögl T., Rief M. Single molecule unzipping of coiled coils: sequence resolved stability profiles. Phys. Rev. Lett. 2006;96:118102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

