Origin of chromosomal translocations in lymphoid cancer
- PMID: 20371343
- PMCID: PMC2874895
- DOI: 10.1016/j.cell.2010.03.016
Origin of chromosomal translocations in lymphoid cancer
Abstract
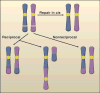
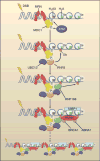
Aberrant fusions between heterologous chromosomes are among the most prevalent cytogenetic abnormalities found in cancer cells. Oncogenic chromosomal translocations provide cells with a proliferative or survival advantage. They may either initiate transformation or be acquired secondarily as a result of genomic instability. Here, we highlight recent advances toward understanding the origin of chromosomal translocations in incipient lymphoid cancers and how tumor-suppressive pathways normally limit the frequency of these aberrant recombination events. Deciphering the mechanisms that mediate chromosomal fusions will open new avenues for developing therapeutic strategies aimed at eliminating lesions that lead to the initiation, maintenance, and progression of cancer.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

References
-
- Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–1135. - PubMed
-
- Bassing CH, Alt FW. Cell cycle. Vol. 3. Georgetown, Tex: 2004. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity; pp. 149–153. - PubMed
-
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

