DNA-binding properties of T4 UvsY recombination mediator protein: polynucleotide wrapping promotes high-affinity binding to single-stranded DNA
- PMID: 20371513
- PMCID: PMC2919719
- DOI: 10.1093/nar/gkq219
DNA-binding properties of T4 UvsY recombination mediator protein: polynucleotide wrapping promotes high-affinity binding to single-stranded DNA
Abstract
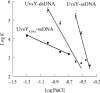
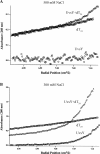
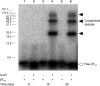
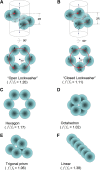
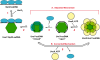
To carry out homologous recombination events in the cell, recombination proteins must be able to recognize and form presynaptic filaments on single-stranded DNA (ssDNA) in the presence of a vast excess of double-stranded DNA (dsDNA). Therefore recombination machineries stringently discriminate between ssDNA and dsDNA lattices. Recent single-molecule studies of bacteriophage T4 recombination proteins revealed that, surprisingly, the UvsY recombination mediator protein binds stronger to stretched dsDNA molecules than to stretched ssDNA. Here, we show that for relaxed DNA lattices, the opposite is true: UvsY exhibits a 1000-fold intrinsic affinity preference for ssDNA over dsDNA at moderate salt concentrations. This finding suggests that UvsY preferentially loads UvsX recombinase onto ssDNA under physiological conditions. The biochemical basis for high-affinity UvsY-ssDNA binding was investigated by hydrodynamic and cross-linking methods. Results show that UvsY forms ring-like hexamers in solution, and that ssDNA binds to multiple subunits within each hexamer, consistent with ssDNA wrapping. The data support a model in which ssDNA wrapping by UvsY protein is important for the selective nucleation of presynaptic filaments on ssDNA versus dsDNA, and for the coordinated transfer of ssDNA from Gp32 (SSB) to UvsY (RMP) to UvsX (recombinase) during filament assembly.
Figures

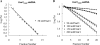
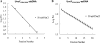
Similar articles
-
Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions.J Biol Chem. 2006 Sep 8;281(36):26308-19. doi: 10.1074/jbc.M604349200. Epub 2006 Jul 7. J Biol Chem. 2006. PMID: 16829679
-
Modulation of T4 gene 32 protein DNA binding activity by the recombination mediator protein UvsY.J Mol Biol. 2008 Jul 25;380(5):799-811. doi: 10.1016/j.jmb.2008.05.039. Epub 2008 May 24. J Mol Biol. 2008. PMID: 18565541 Free PMC article.
-
Mutations in a conserved motif inhibit single-stranded DNA binding and recombination mediator activities of bacteriophage T4 UvsY protein.J Biol Chem. 2004 Feb 13;279(7):6077-86. doi: 10.1074/jbc.M311557200. Epub 2003 Nov 22. J Biol Chem. 2004. PMID: 14634008
-
Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8298-305. doi: 10.1073/pnas.131007498. Proc Natl Acad Sci U S A. 2001. PMID: 11459967 Free PMC article. Review.
-
Biophysical characterization of DNA binding from single molecule force measurements.Phys Life Rev. 2010 Sep;7(3):299-341. doi: 10.1016/j.plrev.2010.06.001. Epub 2010 Jun 4. Phys Life Rev. 2010. PMID: 20576476 Free PMC article. Review.
Cited by
-
Bacteriophage T4 Escapes CRISPR Attack by Minihomology Recombination and Repair.mBio. 2021 Jun 29;12(3):e0136121. doi: 10.1128/mBio.01361-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154416 Free PMC article.
-
Structure and mechanism of the phage T4 recombination mediator protein UvsY.Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):3275-80. doi: 10.1073/pnas.1519154113. Epub 2016 Mar 7. Proc Natl Acad Sci U S A. 2016. PMID: 26951671 Free PMC article.
-
Enzyme-Assisted Nucleic Acid Amplification in Molecular Diagnosis: A Review.Biosensors (Basel). 2023 Jan 19;13(2):160. doi: 10.3390/bios13020160. Biosensors (Basel). 2023. PMID: 36831926 Free PMC article. Review.
-
Assembly and dynamics of the bacteriophage T4 homologous recombination machinery.Virol J. 2010 Dec 3;7:357. doi: 10.1186/1743-422X-7-357. Virol J. 2010. PMID: 21129202 Free PMC article. Review.
-
Presynaptic filament dynamics in homologous recombination and DNA repair.Crit Rev Biochem Mol Biol. 2011 Jun;46(3):240-70. doi: 10.3109/10409238.2011.576007. Crit Rev Biochem Mol Biol. 2011. PMID: 21599536 Free PMC article.
References
-
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. - PubMed
-
- Sung P, Trujillo KM, Van Komen S. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 2000;451:257–275. - PubMed
-
- Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. - PubMed

