Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1
- PMID: 20371601
- PMCID: PMC2878036
- DOI: 10.1074/jbc.M109.082370
Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1
Abstract
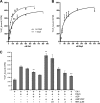
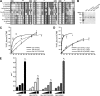
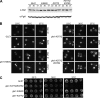
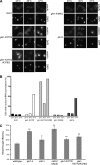
The unidirectional translocation of messenger RNA (mRNA) through the aqueous channel of the nuclear pore complex (NPC) is mediated by interactions between soluble mRNA export factors and distinct binding sites on the NPC. At the cytoplasmic side of the NPC, the conserved mRNA export factors Gle1 and inositol hexakisphosphate (IP(6)) play an essential role in mRNA export by activating the ATPase activity of the DEAD-box protein Dbp5, promoting localized messenger ribonucleoprotein complex remodeling, and ensuring the directionality of the export process. In addition, Dbp5, Gle1, and IP(6) are also required for proper translation termination. However, the specificity of the IP(6)-Gle1 interaction in vivo is unknown. Here, we characterize the biochemical interaction between Gle1 and IP(6) and the relationship to Dbp5 binding and stimulation. We identify Gle1 residues required for IP(6) binding and show that these residues are needed for IP(6)-dependent Dbp5 stimulation in vitro. Furthermore, we demonstrate that Gle1 is the primary target of IP(6) for both mRNA export and translation termination in vivo. In Saccharomyces cerevisiae cells, the IP(6)-binding mutants recapitulate all of the mRNA export and translation termination defects found in mutants depleted of IP(6). We conclude that Gle1 specifically binds IP(6) and that this interaction is required for the full potentiation of Dbp5 ATPase activity during both mRNA export and translation termination.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

