Activation of AP-1 transcription factors differentiates FGF2 and vascular endothelial growth factor regulation of endothelial nitric-oxide synthase expression in placental artery endothelial cells
- PMID: 20371606
- PMCID: PMC2878498
- DOI: 10.1074/jbc.M109.092791
Activation of AP-1 transcription factors differentiates FGF2 and vascular endothelial growth factor regulation of endothelial nitric-oxide synthase expression in placental artery endothelial cells
Abstract
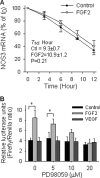
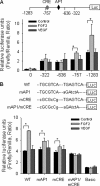
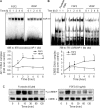
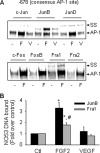
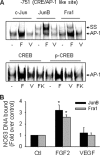
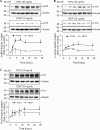
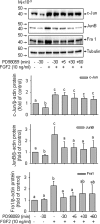
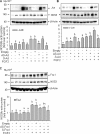
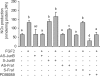
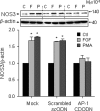
FGF2 (fibroblast growth factor 2), but not vascular endothelial growth factor (VEGF), stimulates sustained activation of ERK2/1 for endothelial NOS3 (nitric-oxide synthase 3) protein expression in ovine fetoplacental artery endothelial cells (oFPAEC). We deciphered herein the downstream signaling of ERK2/1 responsible for NOS3 expression by FGF2 in oFPAEC. FGF2, but not VEGF, increased NOS3 mRNA levels without altering its degradation. FGF2, but not VEGF, trans-activated sheep NOS3 promoter, and this was dependent on ERK2/1 activation. FGF2 did not trans-activate NOS3 promoters with deletions upstream of the consensus AP-1 site (TGAGTC A, -678 to -685). Trans-activation of wild-type NOS3 promoter by FGF2 was significantly inhibited when either the AP-1 or the cAMP-response element (CRE)-like sequence (TGCGTCA, -752 to -758) was mutated and was completely blocked when both were mutated. EMSA analyses showed that FGF2, but not VEGF, stimulated AP-1 and CRE DNA-protein complexes primarily composed of JunB and Fra1. Chromatin immunoprecipitation assays confirmed JunB/Fra1 binding to NOS3 promoter AP-1 and CRE elements in intact cells. FGF2, but not VEGF, stimulated JunB and Fra1 expressions; all preceded NOS3 up-regulation and were inhibited by PD98059. Down-regulation of JunB or Fra-1, but not c-Jun, blocked FGF2 stimulation of NOS3 expression and NO production. AP-1 inhibition suppressed FGF2 stimulation of NOS3 expression in human umbilical vein EC and uterine artery endothelial cells. Thus, FGF2 induction of NOS3 expression is mainly mediated by AP-1-dependent transcription involving JunB and Fra1 up-regulation via sustained ERK2/1 activation in endothelial cells.
Figures
Similar articles
-
Differential activation of multiple signalling pathways dictates eNOS upregulation by FGF2 but not VEGF in placental artery endothelial cells.Placenta. 2008 Aug;29(8):708-17. doi: 10.1016/j.placenta.2008.05.005. Epub 2008 Jun 20. Placenta. 2008. PMID: 18571718 Free PMC article.
-
Transcriptional regulation of endothelial nitric oxide synthase expression in uterine artery endothelial cells by c-Jun/AP-1.Mol Cell Endocrinol. 2007 Dec 15;279(1-2):39-51. doi: 10.1016/j.mce.2007.08.017. Epub 2007 Sep 8. Mol Cell Endocrinol. 2007. PMID: 17933457 Free PMC article.
-
Suppression of protein phosphatase 2 differentially modulates VEGF- and FGF2-induced signaling in ovine fetoplacental artery endothelial cells.Placenta. 2009 Oct;30(10):907-13. doi: 10.1016/j.placenta.2009.07.003. Epub 2009 Aug 18. Placenta. 2009. PMID: 19692121 Free PMC article.
-
Shear stress regulation of nitric oxide production in uterine and placental artery endothelial cells: experimental studies and hemodynamic models of shear stresses on endothelial cells.Int J Dev Biol. 2010;54(2-3):331-9. doi: 10.1387/ijdb.082832bs. Int J Dev Biol. 2010. PMID: 19876820 Free PMC article. Review.
-
Multiple Therapeutic Applications of RBM-007, an Anti-FGF2 Aptamer.Cells. 2021 Jun 28;10(7):1617. doi: 10.3390/cells10071617. Cells. 2021. PMID: 34203430 Free PMC article. Review.
Cited by
-
Regulation of placental angiogenesis.Microcirculation. 2014 Jan;21(1):15-25. doi: 10.1111/micc.12093. Microcirculation. 2014. PMID: 23981199 Free PMC article. Review.
-
Estradiol-17β stimulates H2 S biosynthesis by ER-dependent CBS and CSE transcription in uterine artery smooth muscle cells in vitro.J Cell Physiol. 2019 Jun;234(6):9264-9273. doi: 10.1002/jcp.27606. Epub 2018 Oct 14. J Cell Physiol. 2019. PMID: 30317617 Free PMC article.
-
Signaling regulation of fetoplacental angiogenesis.J Endocrinol. 2012 Mar;212(3):243-55. doi: 10.1530/JOE-11-0296. Epub 2011 Nov 21. J Endocrinol. 2012. PMID: 22106098 Free PMC article. Review.
-
Caveolin-1 orchestrates fibroblast growth factor 2 signaling control of angiogenesis in placental artery endothelial cell caveolae.J Cell Physiol. 2012 Jun;227(6):2480-91. doi: 10.1002/jcp.22984. J Cell Physiol. 2012. PMID: 21830216 Free PMC article.
-
Rac1-dependent intracellular superoxide formation mediates vascular endothelial growth factor-induced placental angiogenesis in vitro.Endocrinology. 2010 Nov;151(11):5315-25. doi: 10.1210/en.2010-0178. Epub 2010 Sep 15. Endocrinology. 2010. PMID: 20844008 Free PMC article.
References
-
- Redman C. W., Sargent I. L. (2005) Science 308, 1592–1594 - PubMed
-
- Borowicz P. P., Arnold D. R., Johnson M. L., Grazul-Bilska A. T., Redmer D. A., Reynolds L. P. (2007) Biol. Reprod. 76, 259–267 - PubMed
-
- Nelson S. H., Steinsland O. S., Wang Y., Yallampalli C., Dong Y. L., Sanchez J. M. (2000) Circ. Res. 87, 406–411 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

