Venom components from Citharischius crawshayi spider (Family Theraphosidae): exploring transcriptome, venomics, and function
- PMID: 20372963
- PMCID: PMC11115658
- DOI: 10.1007/s00018-010-0359-x
Venom components from Citharischius crawshayi spider (Family Theraphosidae): exploring transcriptome, venomics, and function
Abstract
Despite strong efforts, knowledge about the composition of the venom of many spider species remains very limited. This work is the first report of transcriptome and venom analysis of the African spider Citharischius crawshayi. We used combined protocols of transcriptomics, venomics, and biological assays to characterize the venom and genes expressed in venom glands. A cDNA library of the venom glands was constructed and used to generate expressed sequence tags (ESTs). Sequence comparisons from 236 ESTs revealed interesting and unique sequences, corresponding to toxin-like and other components. Mass spectrometrical analysis of venom fractions showed more than 600 molecular masses, some of which showed toxic activity on crickets and modulated sodium currents in DmNa(v)1 and Na(v)1.6 channels as expressed in Xenopus oocytes. Taken together, our results may contribute to a better understanding of the cellular processes involved in the transcriptome and help us to discover new components from spider venom glands with therapeutic potential.
Figures
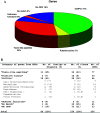


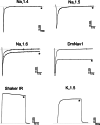
References
-
- Platnick NI (2009) The world spider catalog, version 9.0. In: Merrett P, Cameron HD (eds) American Museum of Natural History, New York. Online at http://research.amnh.org/entomology/spiders/catalog/index.html
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

