Structure and activity of CPNGRC: a modified CD13/APN peptidic homing motif
- PMID: 20374250
- PMCID: PMC2890305
- DOI: 10.1111/j.1747-0285.2010.00974.x
Structure and activity of CPNGRC: a modified CD13/APN peptidic homing motif
Abstract
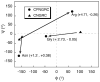
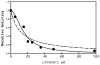
Asn-Gly-Arg peptides have been designed as vehicles for the delivery of chemotherapeutics, magnetic resonance imaging contrast agents, and fluorescence labels to tumor cells, and cardiac angiogenic tissue. Specificity is derived via an interaction with aminopeptidase N, also known as CD13, a cell surface receptor that is highly expressed in angiogenic tissue. Peptides containing the CNGRC homing sequence tethered to a pro-apoptotic peptide sequence have the ability to specifically induce apoptosis in tumor cells. We have now identified a modification to the Asn-Gly-Arg homing sequence motif that improves overall binding affinity to aminopeptidase N. Through the addition of a proline residue, the new peptide with sequence, CPNGRC, inhibits aminopeptidase N proteolytic activity with an IC(50) of 10 microM, a value that is 30-fold lower than that for CNGRC. Both peptides are cyclized via a disulfide bridge between cysteines. Steady-state kinetic experiments suggest that efficient aminopeptidase N inhibition is achieved through the highly cooperative binding of two molecules of CPNGRC. We have used NMR-derived structural constraints for the elucidation of the solution structures CNGRC and CPNGRC. Resulting structures of CNGRC and CPNGRC have significant differences in the backbone torsion angles, which may contribute to the enhanced binding affinity and demonstrated enzyme inhibition by CPNGRC.
Figures








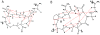
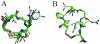

Similar articles
-
NGR Tumor-Homing Peptides: Structural Requirements for Effective APN (CD13) Targeting.Bioconjug Chem. 2016 May 18;27(5):1332-40. doi: 10.1021/acs.bioconjchem.6b00136. Epub 2016 May 2. Bioconjug Chem. 2016. PMID: 27077642
-
A unified mechanism for aminopeptidase N-based tumor cell motility and tumor-homing therapy.J Biol Chem. 2014 Dec 12;289(50):34520-9. doi: 10.1074/jbc.M114.566802. Epub 2014 Oct 29. J Biol Chem. 2014. PMID: 25359769 Free PMC article.
-
The CNGRC-GG-D(KLAKLAK)2 peptide induces a caspase-independent, Ca2+-dependent death in human leukemic myeloid cells by targeting surface aminopeptidase N/CD13.Oncotarget. 2016 Apr 12;7(15):19445-67. doi: 10.18632/oncotarget.6523. Oncotarget. 2016. PMID: 26655501 Free PMC article.
-
Gadolinium-diethylenetriaminepentaacetic acid-cyclo(Cys-Asn-Gly-Arg-Cys)-Gly-Lys-quantum dots.2009 Jan 15 [updated 2009 Mar 19]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2009 Jan 15 [updated 2009 Mar 19]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 20641652 Free Books & Documents. Review.
-
Progress in the development of aminopeptidase N (APN/CD13) inhibitors.Curr Med Chem Anticancer Agents. 2005 May;5(3):281-301. doi: 10.2174/1568011053765949. Curr Med Chem Anticancer Agents. 2005. PMID: 15992355 Review.
Cited by
-
NGR-Based Radiopharmaceuticals for Angiogenesis Imaging: A Preclinical Review.Int J Mol Sci. 2023 Aug 11;24(16):12675. doi: 10.3390/ijms241612675. Int J Mol Sci. 2023. PMID: 37628856 Free PMC article. Review.
-
Microfluidic system for facilitated quantification of nanoparticle accumulation to cells under laminar flow.Ann Biomed Eng. 2013 Jan;41(1):89-99. doi: 10.1007/s10439-012-0634-0. Epub 2012 Aug 2. Ann Biomed Eng. 2013. PMID: 22855121 Free PMC article.
-
Synthesis and evaluation of novel Tc-99m labeled probestin conjugates for imaging APN/CD13 expression in vivo.Bioconjug Chem. 2012 Jan 18;23(1):115-24. doi: 10.1021/bc200546b. Epub 2011 Dec 20. Bioconjug Chem. 2012. PMID: 22148582 Free PMC article.
References
-
- Arap W, Pasqualini R, Ruoslahti E. Cancer Treatment by Targeted Drug Delivery to Tumor Vasculature in a Mouse Model. Science. 1998;279:377–80. - PubMed
-
- Zhang Z, Harada H, Tanabe K, Hatta H, Hiraoka M, Nishimoto S. Aminopeptidase N/CD13 targeting fluorescent probes: synthesis and application to tumor cell imaging. Peptides. 2005;26:2182–7. - PubMed
-
- Colombo G, Curnis F, De Mori GM, Gasparri A, Longoni C, Sacchi A, et al. Structure-activity relationships of linear and cyclic peptides containing the NGR tumor-homing motif. J Biol Chem. 2002;277:47891–7. - PubMed
-
- Corti A, Ponzoni M. Tumor vascular targeting with tumor necrosis factor alpha and chemotherapeutic drugs. Ann N Y Acad Sci. 2004;1028:104–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

