Claudin association with CD81 defines hepatitis C virus entry
- PMID: 20375010
- PMCID: PMC2898367
- DOI: 10.1074/jbc.M110.104836
Claudin association with CD81 defines hepatitis C virus entry
Abstract
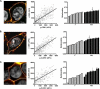
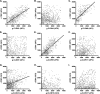
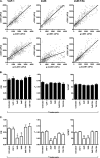
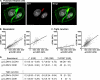
Viruses initiate infection by attaching to molecules or receptors at the cell surface. Hepatitis C virus (HCV) enters cells via a multistep process involving tetraspanin CD81, scavenger receptor class B member I, and the tight junction proteins Claudin-1 and Occludin. CD81 and scavenger receptor class B member I interact with HCV-encoded glycoproteins, suggesting an initial role in mediating virus attachment. In contrast, there are minimal data supporting Claudin-1 association with HCV particles, raising questions as to its role in the virus internalization process. In the present study we demonstrate a relationship between receptor active Claudins and their association and organization with CD81 at the plasma membrane by fluorescence resonance energy transfer and stoichiometric imaging methodologies. Mutation of residues 32 and 48 in the Claudin-1 first extracellular loop ablates CD81 association and HCV receptor activity. Furthermore, mutation of the same residues in the receptor-inactive Claudin-7 molecule enabled CD81 complex formation and virus entry, demonstrating an essential role for Claudin-CD81 complexes in HCV infection. Importantly, Claudin-1 associated with CD81 at the basolateral membrane of polarized HepG2 cells, whereas tight junction-associated pools of Claudin-1 demonstrated a minimal association with CD81. In summary, we demonstrate an essential role for Claudin-CD81 complexes in HCV infection and their localization at the basolateral surface of polarized hepatoma cells, consistent with virus entry into the liver via the sinusoidal blood and association with basal expressed forms of the receptors.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

