Multilevel support vector regression analysis to identify condition-specific regulatory networks
- PMID: 20375112
- PMCID: PMC2872001
- DOI: 10.1093/bioinformatics/btq144
Multilevel support vector regression analysis to identify condition-specific regulatory networks
Abstract
Motivation: The identification of gene regulatory modules is an important yet challenging problem in computational biology. While many computational methods have been proposed to identify regulatory modules, their initial success is largely compromised by a high rate of false positives, especially when applied to human cancer studies. New strategies are needed for reliable regulatory module identification.
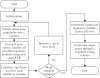
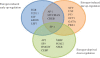
Results: We present a new approach, namely multilevel support vector regression (ml-SVR), to systematically identify condition-specific regulatory modules. The approach is built upon a multilevel analysis strategy designed for suppressing false positive predictions. With this strategy, a regulatory module becomes ever more significant as more relevant gene sets are formed at finer levels. At each level, a two-stage support vector regression (SVR) method is utilized to help reduce false positive predictions by integrating binding motif information and gene expression data; a significant analysis procedure is followed to assess the significance of each regulatory module. To evaluate the effectiveness of the proposed strategy, we first compared the ml-SVR approach with other existing methods on simulation data and yeast cell cycle data. The resulting performance shows that the ml-SVR approach outperforms other methods in the identification of both regulators and their target genes. We then applied our method to breast cancer cell line data to identify condition-specific regulatory modules associated with estrogen treatment. Experimental results show that our method can identify biologically meaningful regulatory modules related to estrogen signaling and action in breast cancer.
Availability and implementation: The ml-SVR MATLAB package can be downloaded at http://www.cbil.ece.vt.edu/software.htm.
Figures


References
-
- Aerts S, et al. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19(Suppl. 2):ii5–ii14. - PubMed
-
- Andrew SD, et al. Sp1 and Sp3 transactivate the RET proto-oncogene promoter. Gene. 2000;256:283–291. - PubMed
-
- Aparicio O, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. 2004 Chapter 17, Unit 17 17. - PubMed
-
- Bar-Joseph Z, et al. Computational discovery of gene modules and regulatory networks. Nat. Biotechnol. 2003;21:1337–1342. - PubMed
-
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 2005;19:833–842. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

