Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus
- PMID: 20375154
- PMCID: PMC2876630
- DOI: 10.1128/JVI.00054-10
Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus
Abstract
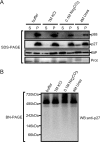
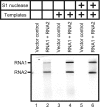
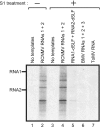
Replication of positive-strand RNA viruses occurs through the assembly of membrane-associated viral RNA replication complexes that include viral replicase proteins, viral RNA templates, and host proteins. Red clover necrotic mosaic virus (RCNMV) is a positive-strand RNA plant virus with a genome consisting of RNA1 and RNA2. The two proteins encoded by RNA1, a 27-kDa protein (p27) and an 88-kDa protein containing an RNA-dependent RNA polymerase (RdRP) motif (p88), are essential for RCNMV RNA replication. To analyze RCNMV RNA replication complexes, we used blue-native polyacrylamide gel electrophoresis (BN/PAGE), which enabled us to analyze detergent-solubilized large membrane protein complexes. p27 and p88 formed a complex of 480 kDa in RCNMV-infected plants. As a result of sucrose gradient sedimentation, the 480-kDa complex cofractionated with both endogenous template-bound and exogenous template-dependent RdRP activities. The amount of the 480-kDa complex corresponded to the activity of exogenous template-dependent RdRP, which produced RNA fragments by specifically recognizing the 3'-terminal core promoter sequences of RCNMV RNAs, but did not correspond to the activity of endogenous template-bound RdRP, which produced genome-sized RNAs without the addition of RNA templates. These results suggest that the 480-kDa complex contributes to template-dependent RdRP activities. We subjected those RdRP complexes to affinity purification and analyzed their components using two-dimensional BN/sodium dodecyl sulfate-PAGE (BN/SDS-PAGE) and mass spectrometry. The 480-kDa complex contained p27, p88, and possible host proteins, and the original affinity-purified RdRP preparation contained HSP70, HSP90, and several ribosomal proteins that were not detected in the 480-kDa complex. A model for the formation of RCNMV RNA replication complexes is proposed.
Figures




References
-
- Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2009. The Red clover necrotic mosaic virus origin of assembly is delimited to the RNA-2 trans-activator. Virology 384:169-178. - PubMed
-
- Bates, H. J., M. Farjah, T. A. M. Osman, and K. W. Buck. 1995. Isolation and characterization of an RNA-dependent RNA polymerase from Nicotiana clevelandii plants infected with red clover necrotic mosaic dianthovirus. J. Gen. Virol. 76:1483-1491. - PubMed
-
- Bonafé, N., M. Gilmore-Hebert, N. L. Folk, M. Azodi, Y. Zhou, and S. K. Chambers. 2005. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 65:3762-3771. - PubMed
-
- Brass, V., E. Bieck, R. Montserret, B. Wolk, J. Hellings, H. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

