Enhancerless cytomegalovirus is capable of establishing a low-level maintenance infection in severely immunodeficient host tissues but fails in exponential growth
- PMID: 20375164
- PMCID: PMC2876661
- DOI: 10.1128/JVI.00419-10
Enhancerless cytomegalovirus is capable of establishing a low-level maintenance infection in severely immunodeficient host tissues but fails in exponential growth
Abstract
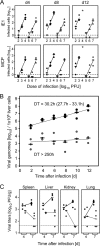
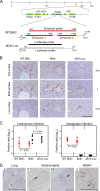
Major immediate-early transcriptional enhancers are genetic control elements that act, through docking with host transcription factors, as a decisive regulatory unit for efficient initiation of the productive virus cycle. Animal models are required for studying the function of enhancers paradigmatically in host organs. Here, we have sought to quantitatively assess the establishment, maintenance, and level of in vivo growth of enhancerless mutants of murine cytomegalovirus in comparison with those of an enhancer-bearing counterpart in models of the immunocompromised or immunologically immature host. Evidence is presented showing that enhancerless viruses are capable of forming restricted foci of infection but fail to grow exponentially.
Figures

Similar articles
-
A temporal gate for viral enhancers to co-opt Toll-like-receptor transcriptional activation pathways upon acute infection.PLoS Pathog. 2015 Apr 9;11(4):e1004737. doi: 10.1371/journal.ppat.1004737. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25856589 Free PMC article.
-
An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis.J Virol. 2003 Mar;77(5):3217-28. doi: 10.1128/jvi.77.5.3217-3228.2003. J Virol. 2003. PMID: 12584345 Free PMC article.
-
Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice.Virology. 1995 May 10;209(1):236-41. doi: 10.1006/viro.1995.1249. Virology. 1995. PMID: 7747475
-
The history of cytomegalovirus and its diseases.Med Microbiol Immunol. 2008 Jun;197(2):65-73. doi: 10.1007/s00430-007-0066-x. Epub 2007 Dec 18. Med Microbiol Immunol. 2008. PMID: 18087722 Review.
-
Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts.Semin Respir Infect. 1995 Dec;10(4):199-208. Semin Respir Infect. 1995. PMID: 8668847 Review.
Cited by
-
Reversible inhibition of murine cytomegalovirus replication by gamma interferon (IFN-γ) in primary macrophages involves a primed type I IFN-signaling subnetwork for full establishment of an immediate-early antiviral state.J Virol. 2011 Oct;85(19):10286-99. doi: 10.1128/JVI.00373-11. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775459 Free PMC article.
-
Principles for studying in vivo attenuation of virus mutants: defining the role of the cytomegalovirus gH/gL/gO complex as a paradigm.Med Microbiol Immunol. 2015 Jun;204(3):295-305. doi: 10.1007/s00430-015-0405-2. Epub 2015 Mar 18. Med Microbiol Immunol. 2015. PMID: 25782576 Review.
-
Viral latency drives 'memory inflation': a unifying hypothesis linking two hallmarks of cytomegalovirus infection.Med Microbiol Immunol. 2012 Nov;201(4):551-66. doi: 10.1007/s00430-012-0273-y. Epub 2012 Sep 19. Med Microbiol Immunol. 2012. PMID: 22991040 Review.
-
A temporal gate for viral enhancers to co-opt Toll-like-receptor transcriptional activation pathways upon acute infection.PLoS Pathog. 2015 Apr 9;11(4):e1004737. doi: 10.1371/journal.ppat.1004737. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25856589 Free PMC article.
-
The activator protein 1 binding motifs within the human cytomegalovirus major immediate-early enhancer are functionally redundant and act in a cooperative manner with the NF-{kappa}B sites during acute infection.J Virol. 2011 Feb;85(4):1732-46. doi: 10.1128/JVI.01713-10. Epub 2010 Nov 24. J Virol. 2011. PMID: 21106746 Free PMC article.
References
-
- Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
-
- Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:60-63. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

