Hepatitis C virus transmission bottlenecks analyzed by deep sequencing
- PMID: 20375170
- PMCID: PMC2876626
- DOI: 10.1128/JVI.02271-09
Hepatitis C virus transmission bottlenecks analyzed by deep sequencing
Abstract
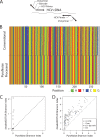
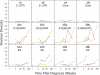
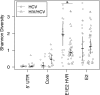
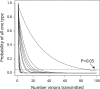
Hepatitis C virus (HCV) replication in infected patients produces large and diverse viral populations, which give rise to drug-resistant and immune escape variants. Here, we analyzed HCV populations during transmission and diversification in longitudinal and cross-sectional samples using 454/Roche pyrosequencing, in total analyzing 174,185 sequence reads. To sample diversity, four locations in the HCV genome were analyzed, ranging from high diversity (the envelope hypervariable region 1 [HVR1]) to almost no diversity (the 5' untranslated region [UTR]). For three longitudinal samples for which early time points were available, we found that only 1 to 4 viral variants were present, suggesting that productive infection was initiated by a very small number of HCV particles. Sequence diversity accumulated subsequently, with the 5' UTR showing almost no diversification while the envelope HVR1 showed >100 variants in some subjects. Calculation of the transmission probability for only a single variant, taking into account the measured population structure within patients, confirmed initial infection by one or a few viral particles. These findings provide the most detailed sequence-based analysis of HCV transmission bottlenecks to date. The analytical methods described here are broadly applicable to studies of viral diversity using deep sequencing.
Figures




References
-
- Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. - PubMed
-
- Anonymous. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. - PubMed
-
- Blackard, J. T., Y. Yang, P. Bordoni, K. E. Sherman, R. T. Chung, and the AIDS Clinical Trials Group 383 Study Team. 2004. Hepatitis C virus (HCV) diversity in HIV-HCV-coinfected subjects initiating highly active antiretroviral therapy. J. Infect. Dis. 189:1472-1481. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

