Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain
- PMID: 20375269
- PMCID: PMC2909294
- DOI: 10.1096/fj.10-154484
Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain
Abstract
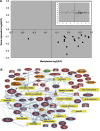
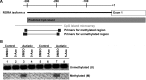
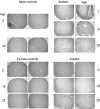
Autism is currently considered a multigene disorder with epigenetic influences. To investigate the contribution of DNA methylation to autism spectrum disorders, we have recently completed large-scale methylation profiling by CpG island microarray analysis of lymphoblastoid cell lines derived from monozygotic twins discordant for diagnosis of autism and their nonautistic siblings. Methylation profiling revealed many candidate genes differentially methylated between discordant MZ twins as well as between both twins and nonautistic siblings. Bioinformatics analysis of the differentially methylated genes demonstrated enrichment for high-level functions including gene transcription, nervous system development, cell death/survival, and other biological processes implicated in autism. The methylation status of 2 of these candidate genes, BCL-2 and retinoic acid-related orphan receptor alpha (RORA), was further confirmed by bisulfite sequencing and methylation-specific PCR, respectively. Immunohistochemical analyses of tissue arrays containing slices of the cerebellum and frontal cortex of autistic and age- and sex-matched control subjects revealed decreased expression of RORA and BCL-2 proteins in the autistic brain. Our data thus confirm the role of epigenetic regulation of gene expression via differential DNA methylation in idiopathic autism, and furthermore link molecular changes in a peripheral cell model with brain pathobiology in autism.
Figures






Similar articles
-
Fetal DNA methylation of autism spectrum disorders candidate genes: association with spontaneous preterm birth.Am J Obstet Gynecol. 2015 Apr;212(4):533.e1-9. doi: 10.1016/j.ajog.2015.02.011. Epub 2015 Feb 14. Am J Obstet Gynecol. 2015. PMID: 25687563
-
A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137.Sci Rep. 2014 Feb 6;4:3994. doi: 10.1038/srep03994. Sci Rep. 2014. PMID: 24500708 Free PMC article.
-
Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism.PLoS One. 2011 Feb 16;6(2):e17116. doi: 10.1371/journal.pone.0017116. PLoS One. 2011. PMID: 21359227 Free PMC article.
-
The genetics of autism.Pediatrics. 2004 May;113(5):e472-86. doi: 10.1542/peds.113.5.e472. Pediatrics. 2004. PMID: 15121991 Review.
-
Is retinoic acid-related orphan receptor-alpha (RORA) a target for gene-environment interactions contributing to autism?Neurotoxicology. 2012 Dec;33(6):1434-1435. doi: 10.1016/j.neuro.2012.07.009. Epub 2012 Aug 8. Neurotoxicology. 2012. PMID: 22967355 Review.
Cited by
-
The expanding genomic landscape of autism: discovering the 'forest' beyond the 'trees'.Future Neurol. 2013 Jan 1;8(1):29-42. doi: 10.2217/fnl.12.83. Future Neurol. 2013. PMID: 23637569 Free PMC article.
-
Malondialdehyde, Bcl-2, superoxide dismutase and glutathione peroxidase may mediate the association of sonic hedgehog protein and oxidative stress in autism.Neurochem Res. 2012 Apr;37(4):899-901. doi: 10.1007/s11064-011-0667-z. Epub 2011 Dec 6. Neurochem Res. 2012. PMID: 22143957 Review.
-
Phenotypic Subtyping and Re-Analysis of Existing Methylation Data from Autistic Probands in Simplex Families Reveal ASD Subtype-Associated Differentially Methylated Genes and Biological Functions.Int J Mol Sci. 2020 Sep 19;21(18):6877. doi: 10.3390/ijms21186877. Int J Mol Sci. 2020. PMID: 32961747 Free PMC article.
-
Association of genes with phenotype in autism spectrum disorder.Aging (Albany NY). 2019 Nov 19;11(22):10742-10770. doi: 10.18632/aging.102473. Epub 2019 Nov 19. Aging (Albany NY). 2019. PMID: 31744938 Free PMC article. Review.
-
An epigenetic biomarker for adult high-functioning autism spectrum disorder.Sci Rep. 2019 Sep 20;9(1):13662. doi: 10.1038/s41598-019-50250-9. Sci Rep. 2019. PMID: 31541176 Free PMC article.
References
-
- Volkmar F. R., Klin A., Siegel B., Szatmari P., Lord C., Campbell M., Freeman B. J., Cicchetti D. V., Rutter M., Kline W. Field trial for autistic disorder in DSM-IV. Am J Psychiatry. 1994;151:1361–1367. - PubMed
-
- Siegel B. Toward DSM-IV: a developmental approach to autistic disorder. Psychiatr Clin N Am. 1991;14:53–68. - PubMed
-
- Bailey A., Le Couteur A., Gottesman I., Bolton P., Simonoff E., Yuzda E., Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. - PubMed
-
- Folstein S., Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. - PubMed
-
- Schanen N. C. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(2):R138–R150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

