Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells
- PMID: 20376339
- PMCID: PMC2848578
- DOI: 10.1371/journal.pone.0009937
Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells
Abstract
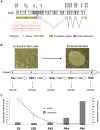
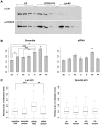
Differentiation of embryonic stem (ES) cells is accompanied by silencing of the Oct-4 gene and de novo DNA methylation of its regulatory region. Previous studies have focused on the requirements for promoter region methylation. We therefore undertook to analyse the progression of DNA methylation of the approximately 2000 base pair regulatory region of Oct-4 in ES cells that are wildtype or deficient for key proteins. We find that de novo methylation is initially seeded at two discrete sites, the proximal enhancer and distal promoter, spreading later to neighboring regions, including the remainder of the promoter. De novo methyltransferases Dnmt3a and Dnmt3b cooperate in the initial targeted stage of de novo methylation. Efficient completion of the pattern requires Dnmt3a and Dnmt1, but not Dnmt3b. Methylation of the Oct-4 promoter depends on the histone H3 lysine 9 methyltransferase G9a, as shown previously, but CpG methylation throughout most of the regulatory region accumulates even in the absence of G9a. Analysis of the Oct-4 regulatory domain as a whole has allowed us to detect targeted de novo methylation and to refine our understanding the roles of key protein components in this process.
Conflict of interest statement
Figures


References
-
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

