Cardiogenic induction of pluripotent stem cells streamlined through a conserved SDF-1/VEGF/BMP2 integrated network
- PMID: 20376342
- PMCID: PMC2848581
- DOI: 10.1371/journal.pone.0009943
Cardiogenic induction of pluripotent stem cells streamlined through a conserved SDF-1/VEGF/BMP2 integrated network
Abstract
Background: Pluripotent stem cells produce tissue-specific lineages through programmed acquisition of sequential gene expression patterns that function as a blueprint for organ formation. As embryonic stem cells respond concomitantly to diverse signaling pathways during differentiation, extraction of a pro-cardiogenic network would offer a roadmap to streamline cardiac progenitor output.
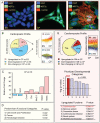
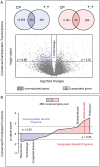
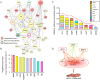
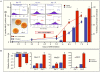
Methods and results: To resolve gene ontology priorities within precursor transcriptomes, cardiogenic subpopulations were here generated according to either growth factor guidance or stage-specific biomarker sorting. Innate expression profiles were independently delineated through unbiased systems biology mapping, and cross-referenced to filter transcriptional noise unmasking a conserved progenitor motif (55 up- and 233 down-regulated genes). The streamlined pool of 288 genes organized into a core biological network that prioritized the "Cardiovascular Development" function. Recursive in silico deconvolution of the cardiogenic neighborhood and associated canonical signaling pathways identified a combination of integrated axes, CXCR4/SDF-1, Flk-1/VEGF and BMP2r/BMP2, predicted to synchronize cardiac specification. In vitro targeting of the resolved triad in embryoid bodies accelerated expression of Nkx2.5, Mef2C and cardiac-MHC, enhanced beating activity, and augmented cardiogenic yield.
Conclusions: Transcriptome-wide dissection of a conserved progenitor profile thus revealed functional highways that coordinate cardiogenic maturation from a pluripotent ground state. Validating the bioinformatics algorithm established a strategy to rationally modulate cell fate, and optimize stem cell-derived cardiogenesis.
Conflict of interest statement
Figures



Similar articles
-
SDF-1-enhanced cardiogenesis requires CXCR4 induction in pluripotent stem cells.J Cardiovasc Transl Res. 2010 Dec;3(6):674-82. doi: 10.1007/s12265-010-9219-1. Epub 2010 Sep 15. J Cardiovasc Transl Res. 2010. PMID: 20842469 Free PMC article.
-
CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells.Stem Cells. 2008 Jun;26(6):1464-73. doi: 10.1634/stemcells.2007-0808. Epub 2008 Mar 27. Stem Cells. 2008. PMID: 18369102
-
Genomic chart guiding embryonic stem cell cardiopoiesis.Genome Biol. 2008 Jan 9;9(1):R6. doi: 10.1186/gb-2008-9-1-r6. Genome Biol. 2008. PMID: 18184438 Free PMC article.
-
Interactome of a cardiopoietic precursor.J Cardiovasc Transl Res. 2008 Jun;1(2):120-6. doi: 10.1007/s12265-008-9019-z. Epub 2008 Jun 3. J Cardiovasc Transl Res. 2008. PMID: 20559906 Review.
-
How to make a cardiomyocyte.Development. 2014 Dec;141(23):4418-31. doi: 10.1242/dev.091538. Epub 2014 Nov 18. Development. 2014. PMID: 25406392 Review.
Cited by
-
Stem cell systems informatics for advanced clinical biodiagnostics: tracing molecular signatures from bench to bedside.Croat Med J. 2013 Aug;54(4):319-29. doi: 10.3325//cmj.2013.54.319. Croat Med J. 2013. PMID: 23986272 Free PMC article. Review.
-
Expression profiles of histone lysine demethylases during cardiomyocyte differentiation of mouse embryonic stem cells.Acta Pharmacol Sin. 2014 Jul;35(7):899-906. doi: 10.1038/aps.2014.40. Acta Pharmacol Sin. 2014. PMID: 24989252 Free PMC article.
-
Challenges for heart disease stem cell therapy.Vasc Health Risk Manag. 2012;8:99-113. doi: 10.2147/VHRM.S25665. Epub 2012 Feb 17. Vasc Health Risk Manag. 2012. PMID: 22399855 Free PMC article. Review.
-
SDF-1-enhanced cardiogenesis requires CXCR4 induction in pluripotent stem cells.J Cardiovasc Transl Res. 2010 Dec;3(6):674-82. doi: 10.1007/s12265-010-9219-1. Epub 2010 Sep 15. J Cardiovasc Transl Res. 2010. PMID: 20842469 Free PMC article.
-
Targeted gene therapy for the treatment of heart failure.Can J Cardiol. 2011 May-Jun;27(3):265-83. doi: 10.1016/j.cjca.2011.02.005. Can J Cardiol. 2011. PMID: 21601767 Free PMC article. Review.
References
-
- Rosenthal N. Prometheus's vulture and the stem-cell promise. N Engl J Med. 2003;349:267–274. - PubMed
-
- Slack JM. Origin of stem cells in organogenesis. Science. 2008;322:1498–1501. - PubMed
-
- Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. - PubMed
-
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. - PubMed
-
- Nelson TJ, Behfar A, Terzic A. Stem cells: biologics for regeneration. Clin Pharmacol Ther. 2008;84:620–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

