Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving
- PMID: 20376398
- PMCID: PMC3128808
- DOI: 10.1039/b926849a
Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving
Abstract
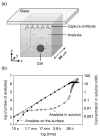
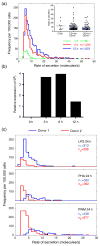
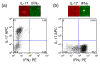
The large diversity of cells that comprise the human immune system requires methods that can resolve the individual contributions of specific subsets to an immunological response. Microengraving is process that uses a dense, elastomeric array of microwells to generate microarrays of proteins secreted from large numbers of individual live cells (approximately 10(4)-10(5) cells/assay). In this paper, we describe an approach based on this technology to quantify the rates of secretion from single immune cells. Numerical simulations of the microengraving process indicated an operating regime between 30 min-4 h that permits quantitative analysis of the rates of secretion. Through experimental validation, we demonstrate that microengraving can provide quantitative measurements of both the frequencies and the distribution in rates of secretion for up to four cytokines simultaneously released from individual viable primary immune cells. The experimental limits of detection ranged from 0.5 to 4 molecules/s for IL-6, IL-17, IFNgamma, IL-2, and TNFalpha. These multidimensional measures resolve the number and intensities of responses by cells exposed to stimuli with greater sensitivity than single-parameter assays for cytokine release. We show that cells from different donors exhibit distinct responses based on both the frequency and magnitude of cytokine secretion when stimulated under different activating conditions. Primary T cells with specific profiles of secretion can also be recovered after microengraving for subsequent expansion in vitro. These examples demonstrate the utility of quantitative, multidimensional profiles of single cells for analyzing the diversity and dynamics of immune responses in vitro and for identifying rare cells from clinical samples.
Figures


References
-
- Pantaleo G, Harari A. Nat Rev Immunol. 2006;6:417–423. - PubMed
-
- Seder RA, Darrah PA, Roederer M. Nat Rev Immunol. 2008;8:247–258. - PubMed
-
- Streeck H, Frahm N, Walker B. Nature Protocols. 2009;4:461–469. - PubMed
-
- Gazagne A, Claret E, Wijdenes J, Yssel H, Bousquet F, Levy E, Vielh P, Scotte F, Goupil TL, Fridman WH, Tartour E. Journal of Immunological Methods. 2003;283:91–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U19AI070352/AI/NIAID NIH HHS/United States
- U19 AI070352/AI/NIAID NIH HHS/United States
- R01 AI091568/AI/NIAID NIH HHS/United States
- F32AI651003/AI/NIAID NIH HHS/United States
- F32 AI065100/AI/NIAID NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- U19 AI050864/AI/NIAID NIH HHS/United States
- P01 AI045757/AI/NIAID NIH HHS/United States
- U19 AI046130/AI/NIAID NIH HHS/United States
- 5U19AI050864-07/AI/NIAID NIH HHS/United States
- U19AI046130/AI/NIAID NIH HHS/United States
- P01AI045757/AI/NIAID NIH HHS/United States
- P01AI039671/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

