Molecular mapping of tumor heterogeneity on clinical tissue specimens with multiplexed quantum dots
- PMID: 20377268
- PMCID: PMC2923482
- DOI: 10.1021/nn100213v
Molecular mapping of tumor heterogeneity on clinical tissue specimens with multiplexed quantum dots
Abstract
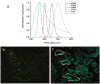
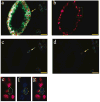
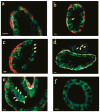
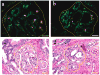
Tumor heterogeneity is one of the most important and challenging problems not only in studying the mechanisms of cancer development but also in developing therapeutics to eradicate cancer cells. Here we report the use of multiplexed quantum dots (QDs) and wavelength-resolved spectral imaging for molecular mapping of tumor heterogeneity on human prostate cancer tissue specimens. By using a panel of just four protein biomarkers (E-cadherin, high-molecular-weight cytokeratin, p63, and alpha-methylacyl CoA racemase), we show that structurally distinct prostate glands and single cancer cells can be detected and characterized within the complex microenvironments of radical prostatectomy and needle biopsy tissue specimens. The results reveal extensive tumor heterogeneity at the molecular, cellular, and architectural levels, allowing direct visualization of human prostate glands undergoing structural transitions from a double layer of basal and luminal cells to a single layer of malignant cells. For clinical diagnostic applications, multiplexed QD mapping provides correlated molecular and morphological information that is not available from traditional tissue staining and molecular profiling methods.
Figures




References
-
- Alivisatos AP. The Use of Nanocrystals in Biological Detection. Nat Biotechnol. 2004;22:47–52. - PubMed
-
- Gao XH, Yang L, Petras JA, Marshall FF, Simons JW, Nie SM. In Vivo Molecular and Cellular Imaging with Quantum Dots. Curr Opin Biotechnol. 2005;16:63–72. - PubMed
-
- Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, Petros JA, O'Regan RM, Yezhelyev MV, Simons JW, et al. Bioconjugated Quantum Dots for Multiplexed and Quantitative Immunohistochemistry. Nat Protoc. 2007;2:1152–1165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

