Introduction of a novel 18S rDNA gene arrangement along with distinct ITS region in the saline water microalga Dunaliella
- PMID: 20377865
- PMCID: PMC2867797
- DOI: 10.1186/1746-1448-6-4
Introduction of a novel 18S rDNA gene arrangement along with distinct ITS region in the saline water microalga Dunaliella
Abstract
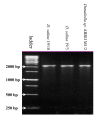
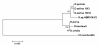
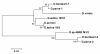
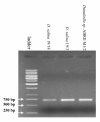
Comparison of 18S rDNA gene sequences is a very promising method for identification and classification of living organisms. Molecular identification and discrimination of different Dunaliella species were carried out based on the size of 18S rDNA gene and, number and position of introns in the gene. Three types of 18S rDNA structure have already been reported: the gene with a size of ~1770 bp lacking any intron, with a size of ~2170 bp consisting one intron near 5' terminus, and with a size of ~2570 bp harbouring two introns near 5' and 3' termini. Hereby, we report a new 18S rDNA gene arrangement in terms of intron localization and nucleotide sequence in a Dunaliella isolated from Iranian salt lakes (ABRIINW-M1/2). PCR amplification with genus-specific primers resulted in production of a ~2170 bp DNA band, which is similar to that of D. salina 18S rDNA gene containing only one intron near 5' terminus. Whilst, sequence composition of the gene revealed the lack of any intron near 5' terminus in our isolate. Furthermore, another alteration was observed due to the presence of a 440 bp DNA fragment near 3' terminus. Accordingly, 18S rDNA gene of the isolate is clearly different from those of D. salina and any other Dunaliella species reported so far. Moreover, analysis of ITS region sequence showed the diversity of this region compared to the previously reported species. 18S rDNA and ITS sequences of our isolate were submitted with accesion numbers of EU678868 and EU927373 in NCBI database, respectively. The optimum growth rate of this isolate occured at the salinity level of 1 M NaCl. The maximum carotenoid content under stress condition of intense light (400 mumol photon m-2 s-1), high salinity (4 M NaCl) and deficiency of nitrate and phosphate nutritions reached to 240 ng/cell after 15 days.
Figures



Similar articles
-
Phenotypic and genetic characterization of Dunaliella (Chlorophyta) from Indian salinas and their diversity.Aquat Biosyst. 2012 Nov 1;8(1):27. doi: 10.1186/2046-9063-8-27. Aquat Biosyst. 2012. PMID: 23114277 Free PMC article.
-
DNA fingerprinting differentiation between beta-carotene hyperproducer strains of Dunaliella from around the world.Saline Syst. 2009 Jun 30;5:5. doi: 10.1186/1746-1448-5-5. Saline Syst. 2009. PMID: 19563682 Free PMC article.
-
Intron polymorphism in small subunit rDNA of Nectria galligena.Microbiology (Reading). 1998 Aug;144 ( Pt 8):2367-2372. doi: 10.1099/00221287-144-8-2367. Microbiology (Reading). 1998. PMID: 9720059
-
Analysis of small and large subunit rDNA introns from several ectomycorrhizal fungi species.PLoS One. 2021 Mar 15;16(3):e0245714. doi: 10.1371/journal.pone.0245714. eCollection 2021. PLoS One. 2021. PMID: 33720962 Free PMC article.
-
Authentication of Curcuma species (Zingiberaceae) based on nuclear 18S rDNA and plastid trnK sequences.Yao Xue Xue Bao. 2010 Jul;45(7):926-33. Yao Xue Xue Bao. 2010. PMID: 20931794
Cited by
-
A simple and inexpensive physical lysis method for DNA and RNA extraction from freshwater microalgae.3 Biotech. 2018 Aug;8(8):354. doi: 10.1007/s13205-018-1381-1. Epub 2018 Aug 1. 3 Biotech. 2018. PMID: 30105179 Free PMC article.
-
Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia.Front Microbiol. 2013 May 13;4:115. doi: 10.3389/fmicb.2013.00115. eCollection 2013. Front Microbiol. 2013. PMID: 23717306 Free PMC article.
-
Phenotypic and genetic characterization of Dunaliella (Chlorophyta) from Indian salinas and their diversity.Aquat Biosyst. 2012 Nov 1;8(1):27. doi: 10.1186/2046-9063-8-27. Aquat Biosyst. 2012. PMID: 23114277 Free PMC article.
-
Efficient and stable transformation of Dunaliella pseudosalina by 3 strains of Agrobacterium tumefaciens.Bioimpacts. 2017;7(4):247-254. doi: 10.15171/bi.2017.29. Epub 2017 Sep 18. Bioimpacts. 2017. PMID: 29435432 Free PMC article.
-
The ecology of Dunaliella in high-salt environments.J Biol Res (Thessalon). 2014 Dec 18;21(1):23. doi: 10.1186/s40709-014-0023-y. eCollection 2014 Dec. J Biol Res (Thessalon). 2014. PMID: 25984505 Free PMC article. Review.
References
-
- Xue L, Pan W, Jiang G, Wang J. Transgenic Dunaliella salina as a bioreactor. United States Patent 7081567. 2001.
-
- Borowitzka MA, Borowitzka LJ. In: Micro-algal Biotechnology. Borowitzka MA, Borowitzka LJ, editor. Cambridge: Cambridge University Press; 1988. Dunaliella; pp. 27–58.
-
- Abatzopoulos TJ, Beardmore JA, Clegg JS, Sorgeloos P. Artemia: Basic and Applied biology. Kluwer Academic Publishers; 2002.
-
- Gómez PI, Gonzaléz MA. Genetic variation among seven strains of Dunaliella salina (Chlorophyta) with industrial potential, based on RAPD banding patterns and on nuclear ITS rDNA sequences. Aquaculture. 2004;233:149–162. doi: 10.1016/j.aquaculture.2003.11.005. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

