The evolution and functional repertoire of translation proteins following the origin of life
- PMID: 20377891
- PMCID: PMC2873265
- DOI: 10.1186/1745-6150-5-15
The evolution and functional repertoire of translation proteins following the origin of life
Abstract
Background: The RNA world hypothesis posits that the earliest genetic system consisted of informational RNA molecules that directed the synthesis of modestly functional RNA molecules. Further evidence suggests that it was within this RNA-based genetic system that life developed the ability to synthesize proteins by translating genetic code. Here we investigate the early development of the translation system through an evolutionary survey of protein architectures associated with modern translation.
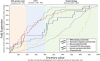
Results: Our analysis reveals a structural expansion of translation proteins immediately following the RNA world and well before the establishment of the DNA genome. Subsequent functional annotation shows that representatives of the ten most ancestral protein architectures are responsible for all of the core protein functions found in modern translation.
Conclusions: We propose that this early robust translation system evolved by virtue of a positive feedback cycle in which the system was able to create increasingly complex proteins to further enhance its own function.
Figures




References
-
- Gilbert W. Origin of life: The RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

