Significantly improved precision of cell migration analysis in time-lapse video microscopy through use of a fully automated tracking system
- PMID: 20377897
- PMCID: PMC2858025
- DOI: 10.1186/1471-2121-11-24
Significantly improved precision of cell migration analysis in time-lapse video microscopy through use of a fully automated tracking system
Abstract
Background: Cell motility is a critical parameter in many physiological as well as pathophysiological processes. In time-lapse video microscopy, manual cell tracking remains the most common method of analyzing migratory behavior of cell populations. In addition to being labor-intensive, this method is susceptible to user-dependent errors regarding the selection of "representative" subsets of cells and manual determination of precise cell positions.
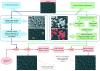
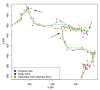
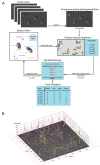
Results: We have quantitatively analyzed these error sources, demonstrating that manual cell tracking of pancreatic cancer cells lead to mis-calculation of migration rates of up to 410%. In order to provide for objective measurements of cell migration rates, we have employed multi-target tracking technologies commonly used in radar applications to develop fully automated cell identification and tracking system suitable for high throughput screening of video sequences of unstained living cells.
Conclusion: We demonstrate that our automatic multi target tracking system identifies cell objects, follows individual cells and computes migration rates with high precision, clearly outperforming manual procedures.
Figures

References
-
- Schneider IC, Haugh JM. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 2006;5(11):1130–1134. - PubMed
-
- Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn MP, Senman B, Junt T. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9(3):282–291. doi: 10.1038/ni1559. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

