Identification of copy number variations and common deletion polymorphisms in cattle
- PMID: 20377913
- PMCID: PMC2859865
- DOI: 10.1186/1471-2164-11-232
Identification of copy number variations and common deletion polymorphisms in cattle
Abstract
Background: Recently, the discovery of copy number variation (CNV) led researchers to think that there are more variations of genomic DNA than initially believed. Moreover, a certain CNV region has been found to be associated with the onset of diseases. Therefore, CNV is now known as an important genomic variation in biological mechanisms. However, most CNV studies have only involved the human genome. The study of CNV involving other animals, including cattle, is severely lacking.
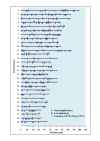
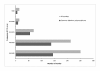
Results: In our study of cattle, we used Illumina BovineSNP50 BeadChip (54,001 markers) to obtain each marker's signal intensity (Log R ratio) and allelic intensity (B allele frequency), which led to our discovery of 855 bovine CNVs from 265 cows. For these animals, the average number of CNVs was 3.2, average size was 149.8 kb, and median size was 171.5 kb. Taking into consideration some overlapping regions among the identified bovine CNVs, 368 unique CNV regions were detected. Among them, there were 76 common CNVRs with > 1% CNV frequency. Together, these CNVRs contained 538 genes. Heritability errors of 156 bovine pedigrees and comparative pairwise analyses were analyzed to detect 448 common deletion polymorphisms. Identified variations in this study were successfully validated using visual examination of the genoplot image, Mendelian inconsistency, another CNV identification program, and quantitative PCR.
Conclusions: In this study, we describe a map of bovine CNVs and provide important resources for future bovine genome research. This result will contribute to animal breeding and protection from diseases with the aid of genomic information.
Figures


References
-
- Casas E, White SN, Riley DG, Smith TP, Brenneman RA, Olson TA, Johnson DD, Coleman SW, Bennett GL, Chase CC Jr. Assessment of single nucleotide polymorphisms in genes residing on chromosomes 14 and 29 for association with carcass composition traits in Bos indicus cattle. J Anim Sci. 2005;83(1):13–19. - PubMed
-
- Gan QF, Zhang LP, Li JY, Hou GY, Li HD, Gao X, Ren HY, Chen JB, Xu SZ. Association analysis of thyroglobulin gene variants with carcass and meat quality traits in beef cattle. J Appl Genet. 2008;49(3):251–255. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

