The longin SNARE VAMP7/TI-VAMP adopts a closed conformation
- PMID: 20378544
- PMCID: PMC2878558
- DOI: 10.1074/jbc.M110.120972
The longin SNARE VAMP7/TI-VAMP adopts a closed conformation
Abstract
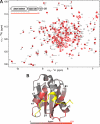
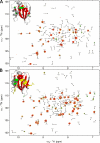
SNARE protein complexes are key mediators of exocytosis by juxtaposing opposing membranes, leading to membrane fusion. SNAREs generally consist of one or two core domains that can form a four-helix bundle with other SNARE core domains. Some SNAREs, such as syntaxin target-SNAREs and longin vesicular-SNAREs, have independent, folded N-terminal domains that can interact with their respective SNARE core domains and thereby affect the kinetics of SNARE complex formation. This autoinhibition mechanism is believed to regulate the role of the longin VAMP7/TI-VAMP in neuronal morphogenesis. Here we use nuclear magnetic resonance spectroscopy to study the longin-SNARE core domain interaction for VAMP7. Using complete backbone resonance assignments, chemical shift perturbations analysis, and hydrogen/deuterium exchange experiments, we conclusively show that VAMP7 adopts a preferentially closed conformation in solution. Taken together, the closed conformation of longins is conserved, in contrast to the syntaxin family of SNAREs for which mixtures of open and closed states have been observed. This may indicate different regulatory mechanisms for SNARE complexes containing syntaxins and longins, respectively.
Figures


Similar articles
-
Alternative splicing of the human gene SYBL1 modulates protein domain architecture of Longin VAMP7/TI-VAMP, showing both non-SNARE and synaptobrevin-like isoforms.BMC Mol Biol. 2011 May 24;12:26. doi: 10.1186/1471-2199-12-26. BMC Mol Biol. 2011. PMID: 21609427 Free PMC article.
-
Increased activity of the vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor TI-VAMP/VAMP7 by tyrosine phosphorylation in the Longin domain.J Biol Chem. 2013 Apr 26;288(17):11960-72. doi: 10.1074/jbc.M112.415075. Epub 2013 Mar 7. J Biol Chem. 2013. PMID: 23471971 Free PMC article.
-
The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly.J Biol Chem. 2002 Sep 27;277(39):36449-56. doi: 10.1074/jbc.M204369200. Epub 2002 Jul 11. J Biol Chem. 2002. PMID: 12114520
-
Structure and function of longin SNAREs.J Cell Sci. 2015 Dec 1;128(23):4263-72. doi: 10.1242/jcs.178574. Epub 2015 Nov 13. J Cell Sci. 2015. PMID: 26567219 Review.
-
Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators.Trends Biochem Sci. 2004 Dec;29(12):682-8. doi: 10.1016/j.tibs.2004.10.002. Trends Biochem Sci. 2004. PMID: 15544955 Review.
Cited by
-
The STX17-SNAP47-VAMP7/VAMP8 complex is the default SNARE complex mediating autophagosome-lysosome fusion.Cell Res. 2024 Feb;34(2):151-168. doi: 10.1038/s41422-023-00916-x. Epub 2024 Jan 5. Cell Res. 2024. PMID: 38182888 Free PMC article.
-
Uncoupling the functions of CALM in VAMP sorting and clathrin-coated pit formation.PLoS One. 2013 May 31;8(5):e64514. doi: 10.1371/journal.pone.0064514. Print 2013. PLoS One. 2013. PMID: 23741335 Free PMC article.
-
VAMP7j: A Splice Variant of Human VAMP7 That Modulates Neurite Outgrowth by Regulating L1CAM Transport to the Plasma Membrane.Int J Mol Sci. 2023 Dec 10;24(24):17326. doi: 10.3390/ijms242417326. Int J Mol Sci. 2023. PMID: 38139155 Free PMC article.
-
The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation.Nat Struct Mol Biol. 2012 Dec;19(12):1300-9. doi: 10.1038/nsmb.2414. Epub 2012 Oct 28. Nat Struct Mol Biol. 2012. PMID: 23104059 Free PMC article.
-
A BLOC-1-AP-3 super-complex sorts a cis-SNARE complex into endosome-derived tubular transport carriers.J Cell Biol. 2021 Jul 5;220(7):e202005173. doi: 10.1083/jcb.202005173. J Cell Biol. 2021. PMID: 33886957 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

