Characterization of the transcriptional regulator Rv3124 of Mycobacterium tuberculosis identifies it as a positive regulator of molybdopterin biosynthesis and defines the functional consequences of a non-synonymous SNP in the Mycobacterium bovis BCG orthologue
- PMID: 20378651
- PMCID: PMC3068679
- DOI: 10.1099/mic.0.037200-0
Characterization of the transcriptional regulator Rv3124 of Mycobacterium tuberculosis identifies it as a positive regulator of molybdopterin biosynthesis and defines the functional consequences of a non-synonymous SNP in the Mycobacterium bovis BCG orthologue
Abstract
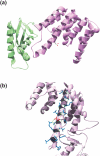
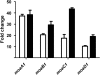
A number of single-nucleotide polymorphisms (SNPs) have been identified in the genome of Mycobacterium bovis BCG Pasteur compared with the sequenced strain M. bovis 2122/97. The functional consequences of many of these mutations remain to be described; however, mutations in genes encoding regulators may be particularly relevant to global phenotypic changes such as loss of virulence, since alteration of a regulator's function will affect the expression of a wide range of genes. One such SNP falls in bcg3145, encoding a member of the AfsR/DnrI/SARP class of global transcriptional regulators, that replaces a highly conserved glutamic acid residue at position 159 (E159G) with glycine in a tetratricopeptide repeat (TPR) located in the bacterial transcriptional activation (BTA) domain of BCG3145. TPR domains are associated with protein-protein interactions, and a conserved core (helices T1-T7) of the BTA domain seems to be required for proper function of SARP-family proteins. Structural modelling predicted that the E159G mutation perturbs the third alpha-helix of the BTA domain and could therefore have functional consequences. The E159G SNP was found to be present in all BCG strains, but absent from virulent M. bovis and Mycobacterium tuberculosis strains. By overexpressing BCG3145 and Rv3124 in BCG and H37Rv and monitoring transcriptome changes using microarrays, we determined that BCG3145/Rv3124 acts as a positive transcriptional regulator of the molybdopterin biosynthesis moa1 locus, and we suggest that rv3124 be renamed moaR1. The SNP in bcg3145 was found to have a subtle effect on the activity of MoaR1, suggesting that this mutation is not a key event in the attenuation of BCG.
Figures


Similar articles
-
Single nucleotide polymorphisms that cause structural changes in the cyclic AMP receptor protein transcriptional regulator of the tuberculosis vaccine strain Mycobacterium bovis BCG alter global gene expression without attenuating growth.Infect Immun. 2008 May;76(5):2227-34. doi: 10.1128/IAI.01410-07. Epub 2008 Mar 10. Infect Immun. 2008. PMID: 18332206 Free PMC article.
-
Point mutations in the DNA- and cNMP-binding domains of the homologue of the cAMP receptor protein (CRP) in Mycobacterium bovis BCG: implications for the inactivation of a global regulator and strain attenuation.Microbiology (Reading). 2005 Feb;151(Pt 2):547-556. doi: 10.1099/mic.0.27444-0. Microbiology (Reading). 2005. PMID: 15699203
-
Differential carriage of virulence-associated loci in the New Zealand Rangipo outbreak strain of Mycobacterium tuberculosis.Infect Dis (Lond). 2017 Sep;49(9):680-688. doi: 10.1080/23744235.2017.1330553. Epub 2017 May 23. Infect Dis (Lond). 2017. PMID: 28535727
-
Genomics of Mycobacterium bovis.Tuberculosis (Edinb). 2001;81(1-2):157-63. doi: 10.1054/tube.2000.0269. Tuberculosis (Edinb). 2001. PMID: 11463237 Review.
-
Long range transcriptional control of virulence critical genes in Mycobacterium tuberculosis by nucleoid-associated proteins?Virulence. 2012 Jul 1;3(4):408-10. doi: 10.4161/viru.20918. Epub 2012 Jun 22. Virulence. 2012. PMID: 22722242 Free PMC article. Review. No abstract available.
Cited by
-
Horizontal acquisition of a hypoxia-responsive molybdenum cofactor biosynthesis pathway contributed to Mycobacterium tuberculosis pathoadaptation.PLoS Pathog. 2017 Nov 27;13(11):e1006752. doi: 10.1371/journal.ppat.1006752. eCollection 2017 Nov. PLoS Pathog. 2017. PMID: 29176894 Free PMC article.
-
Whole transcriptomic and proteomic analyses of an isogenic M. tuberculosis clinical strain with a naturally occurring 15 Kb genomic deletion.PLoS One. 2017 Jun 26;12(6):e0179996. doi: 10.1371/journal.pone.0179996. eCollection 2017. PLoS One. 2017. PMID: 28650996 Free PMC article.
-
Commonalities of Mycobacterium tuberculosis Transcriptomes in Response to Defined Persisting Macrophage Stresses.Front Immunol. 2022 Jul 1;13:909904. doi: 10.3389/fimmu.2022.909904. eCollection 2022. Front Immunol. 2022. PMID: 35844560 Free PMC article.
-
Database resources for the tuberculosis community.Tuberculosis (Edinb). 2013 Jan;93(1):12-7. doi: 10.1016/j.tube.2012.11.003. Epub 2013 Jan 17. Tuberculosis (Edinb). 2013. PMID: 23332401 Free PMC article. Review.
-
HigA2 (Rv2021c) Is a Transcriptional Regulator with Multiple Regulatory Targets in Mycobacterium tuberculosis.Microorganisms. 2024 Jun 20;12(6):1244. doi: 10.3390/microorganisms12061244. Microorganisms. 2024. PMID: 38930627 Free PMC article.
References
-
- Baker, K. P. & Boxer, D. H. (1991). Regulation of the chlA locus of Escherichia coli K12: involvement of molybdenum cofactor. Mol Microbiol 5, 901–907. - PubMed
-
- Bardarov, S., Bardarov, S., Jr, Pavelka, M. S., Jr, Sambandamurthy, V., Larsen, M., Tufariello, J., Chan, J., Hatfull, G. & Jacobs, W. R., Jr (2002). Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017. - PubMed
-
- Behr, M. A., Wilson, M. A., Gill, W. P., Salamon, H., Schoolnik, G. K., Rane, S. & Small, P. M. (1999). Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284, 1520–1523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

