Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities
- PMID: 20378718
- PMCID: PMC2877582
- DOI: 10.1101/gr.100552.109
Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities
Abstract
The genetic code-the binding specificity of all transfer-RNAs--defines how protein primary structure is determined by DNA sequence. DNA also dictates when and where proteins are expressed, and this information is encoded in a pattern of specific sequence motifs that are recognized by transcription factors. However, the DNA-binding specificity is only known for a small fraction of the approximately 1400 human transcription factors (TFs). We describe here a high-throughput method for analyzing transcription factor binding specificity that is based on systematic evolution of ligands by exponential enrichment (SELEX) and massively parallel sequencing. The method is optimized for analysis of large numbers of TFs in parallel through the use of affinity-tagged proteins, barcoded selection oligonucleotides, and multiplexed sequencing. Data are analyzed by a new bioinformatic platform that uses the hundreds of thousands of sequencing reads obtained to control the quality of the experiments and to generate binding motifs for the TFs. The described technology allows higher throughput and identification of much longer binding profiles than current microarray-based methods. In addition, as our method is based on proteins expressed in mammalian cells, it can also be used to characterize DNA-binding preferences of full-length proteins or proteins requiring post-translational modifications. We validate the method by determining binding specificities of 14 different classes of TFs and by confirming the specificities for NFATC1 and RFX3 using ChIP-seq. Our results reveal unexpected dimeric modes of binding for several factors that were thought to preferentially bind DNA as monomers.
Figures



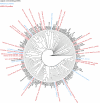

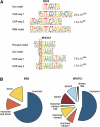
References
-
- Bailey TL, Elkan C 1995. The value of prior knowledge in discovering motifs with MEME. Technical Report CS95-413. Department of Computer Science, University of California, San Diego - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
