Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding
- PMID: 20378774
- PMCID: PMC3008766
- DOI: 10.1126/science.1186176
Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding
Abstract
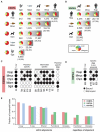
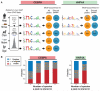
Transcription factors (TFs) direct gene expression by binding to DNA regulatory regions. To explore the evolution of gene regulation, we used chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) to determine experimentally the genome-wide occupancy of two TFs, CCAAT/enhancer-binding protein alpha and hepatocyte nuclear factor 4 alpha, in the livers of five vertebrates. Although each TF displays highly conserved DNA binding preferences, most binding is species-specific, and aligned binding events present in all five species are rare. Regions near genes with expression levels that are dependent on a TF are often bound by the TF in multiple species yet show no enhanced DNA sequence constraint. Binding divergence between species can be largely explained by sequence changes to the bound motifs. Among the binding events lost in one lineage, only half are recovered by another binding event within 10 kilobases. Our results reveal large interspecies differences in transcriptional regulation and provide insight into regulatory evolution.
Figures


Comment in
-
Dramatic changes in transcription factor binding over evolutionary time.Genome Biol. 2010;11(6):122. doi: 10.1186/gb-2010-11-6-122. Epub 2010 Jun 1. Genome Biol. 2010. PMID: 20519030 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

