NetCTLpan: pan-specific MHC class I pathway epitope predictions
- PMID: 20379710
- PMCID: PMC2875469
- DOI: 10.1007/s00251-010-0441-4
NetCTLpan: pan-specific MHC class I pathway epitope predictions
Abstract
Reliable predictions of immunogenic peptides are essential in rational vaccine design and can minimize the experimental effort needed to identify epitopes. In this work, we describe a pan-specific major histocompatibility complex (MHC) class I epitope predictor, NetCTLpan. The method integrates predictions of proteasomal cleavage, transporter associated with antigen processing (TAP) transport efficiency, and MHC class I binding affinity into a MHC class I pathway likelihood score and is an improved and extended version of NetCTL. The NetCTLpan method performs predictions for all MHC class I molecules with known protein sequence and allows predictions for 8-, 9-, 10-, and 11-mer peptides. In order to meet the need for a low false positive rate, the method is optimized to achieve high specificity. The method was trained and validated on large datasets of experimentally identified MHC class I ligands and cytotoxic T lymphocyte (CTL) epitopes. It has been reported that MHC molecules are differentially dependent on TAP transport and proteasomal cleavage. Here, we did not find any consistent signs of such MHC dependencies, and the NetCTLpan method is implemented with fixed weights for proteasomal cleavage and TAP transport for all MHC molecules. The predictive performance of the NetCTLpan method was shown to outperform other state-of-the-art CTL epitope prediction methods. Our results further confirm the importance of using full-type human leukocyte antigen restriction information when identifying MHC class I epitopes. Using the NetCTLpan method, the experimental effort to identify 90% of new epitopes can be reduced by 15% and 40%, respectively, when compared to the NetMHCpan and NetCTL methods. The method and benchmark datasets are available at http://www.cbs.dtu.dk/services/NetCTLpan/.
Figures
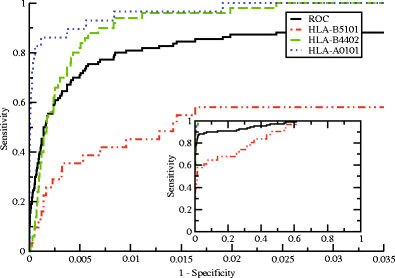

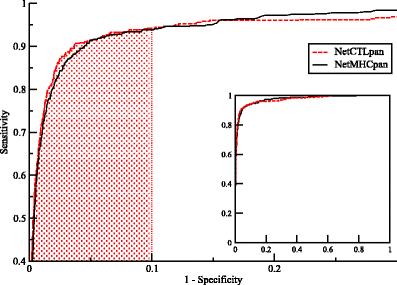
Similar articles
-
An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions.Eur J Immunol. 2005 Aug;35(8):2295-303. doi: 10.1002/eji.200425811. Eur J Immunol. 2005. PMID: 15997466
-
Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction.BMC Bioinformatics. 2007 Oct 31;8:424. doi: 10.1186/1471-2105-8-424. BMC Bioinformatics. 2007. PMID: 17973982 Free PMC article.
-
Cytotoxic T lymphocyte epitopes of HIV-1 Nef: Generation of multiple definitive major histocompatibility complex class I ligands by proteasomes.J Exp Med. 2000 Jan 17;191(2):239-52. doi: 10.1084/jem.191.2.239. J Exp Med. 2000. PMID: 10637269 Free PMC article.
-
The specificity of proteasomes: impact on MHC class I processing and presentation of antigens.Immunol Rev. 1999 Dec;172:29-48. doi: 10.1111/j.1600-065x.1999.tb01354.x. Immunol Rev. 1999. PMID: 10631935 Review.
-
Function of the transport complex TAP in cellular immune recognition.Biochim Biophys Acta. 1999 Dec 6;1461(2):405-19. doi: 10.1016/s0005-2736(99)00171-6. Biochim Biophys Acta. 1999. PMID: 10581370 Review.
Cited by
-
Immunoinformatics Identification of the Conserved and Cross-Reactive T-Cell Epitopes of SARS-CoV-2 with Human Common Cold Coronaviruses, SARS-CoV, MERS-CoV and Live Attenuated Vaccines Presented by HLA Alleles of Indonesian Population.Viruses. 2022 Oct 24;14(11):2328. doi: 10.3390/v14112328. Viruses. 2022. PMID: 36366426 Free PMC article.
-
Development of innovative multi-epitope mRNA vaccine against central nervous system tuberculosis using in silico approaches.PLoS One. 2024 Sep 6;19(9):e0307877. doi: 10.1371/journal.pone.0307877. eCollection 2024. PLoS One. 2024. PMID: 39240891 Free PMC article.
-
Machine Learning for Cancer Immunotherapies Based on Epitope Recognition by T Cell Receptors.Front Genet. 2019 Nov 19;10:1141. doi: 10.3389/fgene.2019.01141. eCollection 2019. Front Genet. 2019. PMID: 31798635 Free PMC article. Review.
-
pTuneos: prioritizing tumor neoantigens from next-generation sequencing data.Genome Med. 2019 Oct 30;11(1):67. doi: 10.1186/s13073-019-0679-x. Genome Med. 2019. PMID: 31666118 Free PMC article.
-
Recent trends in next generation immunoinformatics harnessed for universal coronavirus vaccine design.Pathog Glob Health. 2023 Mar;117(2):134-151. doi: 10.1080/20477724.2022.2072456. Epub 2022 May 12. Pathog Glob Health. 2023. PMID: 35550001 Free PMC article. Review.
References
-
- Anderson KS, Alexander J, Wei M, Cresswell P. Intracellular transport of class I MHC molecules in antigen processing mutant cell lines. J Immunol. 1993;151:3407–3419. - PubMed
-
- Brusic V, van Endert P, Zeleznikow J, Daniel S, Hammer J, Petrovsky N. A neural network model approach to the study of human TAP transporter. In Silico Biol. 1999;1:109–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

