ADAR editing in double-stranded UTRs and other noncoding RNA sequences
- PMID: 20382028
- PMCID: PMC2897959
- DOI: 10.1016/j.tibs.2010.02.008
ADAR editing in double-stranded UTRs and other noncoding RNA sequences
Abstract
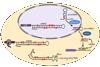
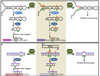
ADARs are a family of enzymes, present in all animals, that convert adenosine to inosine within double-stranded RNA (dsRNA). Inosine and adenosine have different base-pairing properties, and thus, editing alters RNA structure, coding potential and splicing patterns. The first ADAR substrates identified were edited in codons, and ADARs were presumed to function primarily in proteome diversification. Although this is an important function of ADARs, especially in the nervous system, editing in coding sequences is rare compared to editing in noncoding sequences. Introns and untranslated regions of mRNA are the primary noncoding targets, but editing also occurs in small RNAs, such as miRNAs. Although the role of editing in noncoding sequences remains unclear, ongoing research suggests functions in the regulation of a variety of post-transcriptional processes.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. - PubMed
-
- Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem. Rev. 2006;106:3397–3411. - PubMed
-
- Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

